
Human leukocyte antigen (HLA) and cancer immunotherapy: HLA-dependent and -independent adoptive immunotherapies
Cancer immunotherapy exploits the immune system to fight cancers. Although immunotherapy has been an important component of cancer treatment for decades, it does not attract too much attention until the past ten years, especially when the Nobel Prize in Physiology or Medicine was awarded for the discovery of cytotoxic T-lymphocyte-associated protein (CTLA-4) and programmed cell death protein 1/programmed cell death protein ligand 1 (PD-1/PD-L1) (1) in 2018. Nowadays, immune checkpoint inhibitors are used to treat various cancers, including the first line treatment of advanced non-small cell lung cancer (NSCLC), melanoma and renal cell carcinoma. In addition to immune checkpoint inhibitors, adoptive cell therapies and tumor vaccines are also common cancer immunotherapies. The basic mechanism for these cancer immunotherapies is that T cells exert an immune function via recognizing tumor antigens presented by the major histocompatibility complex (MHC) on the membranes of tumor cells (2,3).
MHC is a group of polymorphic genes expressed in nearly all the vertebrates, which determines histocompatibility between different individuals. MHC was first discovered during the first decade of the 20th century because of tumor rejection between genetically distinct mice (4). Human MHC is also known as human leukocyte antigen (HLA).
HLA class I molecules are expressed on most cell types including tumor cells in human, presenting endogenous antigens to the immune system. HLA class II molecules are mainly expressed by antigen-presenting cells (APCs), presenting exogenous antigens to T helper cells. Both HLA class I and class II molecules show high polymorphism, which means that they have many different alleles among human populations. This is why HLA mismatch between donors and recipients is the primary cause of transplant rejection (5). According to IMGT/HLA database, more than 12,000 alleles were identified as HLA class I genes (6). Evolutionarily, the diversity of HLA molecules ensures that the immune system could recognize as many antigens as possible and help us to defend various pathogens. Interestingly, while investigating the effect of HLA class I divergence on the efficacy of immune checkpoint inhibitor treatment for cancer (7), Chowell et al. found that greater sequence divergence of an HLA-I genotype is associated with higher diversity of self, tumor and viral immunopeptidomes. Furthermore, patients with high HLA-I divergence show better responses to immune checkpoint inhibitors than patients with low HLA-I divergence. These findings suggested that HLA polymorphism is critical for us to fight cancer.
Antigen presentation by HLA
HLAs presents tumor antigens to T cells to facilitate the immune system to recognize tumor cells. The process by which HLA molecules bind antigen peptides and present them on the cell membrane is called antigen presentation. Here we summarized three different pathways of antigen presentation (Figure 1). All of these processes occur during tumor development and mediate responses to immunotherapies.
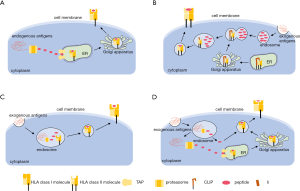
The peptide binding groove of HLA class I molecules is closed at both ends by conserved tyrosine residues, which usually restricts the size of bound peptides to 8–10 residues (8,9). Endogenous antigens are degraded by proteasomes into short peptides in the cytoplasm, and these short peptides are transferred from the cytoplasm to the endoplasmic reticulum (ER) lumen through the antigen processing-related transporter protein (TAP) (Figure 1A). In the ER, empty class I molecules waiting for peptide loading is retained by a series of chaperones including a dedicated chaperone tapasin (also called TAP-binding protein, TAPBP) in the peptide-loading complex. With the help of several chaperones, appropriate peptides then bind to class I molecules and the peptide-HLA complexes become stabilized. The stable complexes continue to move along with the ER, pass through the Golgi apparatus, and finally reach the surface of the cell membrane. In addition to tapasin, a second MHC class I-specific chaperone, the tapasin-related protein TAPBP-R was identified in 2013 (10). Both tapasin and TAPBP-R function as a peptide exchange catalyst and a quality control checkpoint. They ensure MHC class I molecules are loaded with high-affinity peptides and prolong cell surface presence of MHC class I molecules (11).
HLA class II molecules usually bind peptides with 13–25 residues in length according to their open binding grooves (12). Exogenous antigens are taken up by endocytosis or phagocytosis and cleaved into peptides in endosome. HLA class II molecules are synthesized in ER where they pair with a third chain, the invariant chain (Ii) (Figure 1B). This interaction prevents the loading of endogenous peptides to the MHC class II cleft. Ii also guides HLA class II through the cells to a late endosomal MHC class II compartment (MIIC). In MIIC, Ii is proteolytically cleaved into a short peptide called class II-associated Ii peptide (CLIP), which continues to block the binding of peptides to the class II cleft. The CLIP is exchanged for an antigenic peptide with the help of chaperones such as HLA-DM and HLA-DO. Like tapasin and TAPBP-R in the HLA I process, HLA-DM and HLA-DO also shape the peptide repertoire of HLA II that is ultimately presented on the cell surface of CD4+ T cells. Interestingly, Yamashita et al. recently found that HLA-DP molecules with β-chains encoding Gly84 (DP84Gly) do not bind Ii through the CLIP region, nor present CLIP. DP84Gly uniquely exploits both class I and II antigen pathways to present both endogenous and exogenous peptides (13).
Certain APCs such as dendritic cells (DCs) have the ability to process and present exogenous antigens with HLA class I molecules. This process is called cross-presentation. During cross-presentation, extracellular proteins or cell debris are internalized by DCs through endocytosis or phagocytosis and further degraded into peptides and presented onto HLA class I molecules (14,15). There are mainly two pathways that have been reported for cross-presentation: the vacuolar pathway and the cytosolic pathway (Figure 1C). In the vacuolar pathway, an extracellular antigen is taken by DCs into the endosome. The antigen is degraded by proteasome and then the derived peptides are loaded onto HLA class I molecules directly in the endosome. In the cytosolic pathway, like the vacuolar pathway, the extracellular antigen is also internalized into the endosome. However, the antigen may be degraded inside the endosome or transported out of the endosome and then degraded in the cytoplasm. The derived peptides can either be transported by the TAP transporter into the ER, or back into the endosome for loading onto HLA class I molecules. Both pathways may occur in DCs during cross-presentation. The importance of cross-presentation is that it allows DCs to acquire antigens from other pathogen infected cells or cancer cells in the periphery and then report their presence to naive CD8+ T cells in lymphoid organs (16).
HLA-dependent immunotherapy
CD8+ cytotoxic T lymphocytes (CTLs)-based immunotherapy
CD8+ cytotoxic T lymphocytes represent a crucial component of the adaptive immune system against tumors. Most cytotoxic T cells express T cell receptors (TCRs) that can recognize a specific antigen. After TCR binding to peptide-HLA complex expressed by tumor cells, CTLs are activated, accumulate at the tumor site, and release effectors to attack tumor cells. CTLs exert specific killing effects on tumor cells via two major pathways: (I) releasing cytotoxic substances such as perforin, granzymes, and cytokines to kill tumor cells; (II) activating death receptor pathway, e.g., FasL, expressed on the surface of T cells, binds to the death receptor Fas on the surface of tumor cells, to initiate apoptosis-related signals, which lead to tumor cell apoptosis.
Extensive evidence has suggested that adoptive transfer of CTLs could harness the cellular immune system and lead to the killing of tumor cells both in animal models and cancer patients (17). Over the past decades, many studies have transferred in vitro expanded tumor-infiltrating lymphocytes (TILs) back into patient donors and showed promising outcomes. The first one was reported by Rosenberg et al. in 1988 to treat patients with metastatic melanoma (18). Since then, TIL therapy has shown satisfactory efficacy in advanced melanoma. Favorable objective response rates up to 72% were reached with TIL therapy in several consecutive clinical trials, in which 10–20% of treated patients reached a complete remission and 40% of patients achieved durable clinical responses (19). In addition to melanoma, investigators also isolated TILs from other solid tumors such as renal cell, breast and cervical cancer. However, the tumor reactivity of TILs from these other tumors is usually lower when compared to melanoma (19). Although TIL treatment is effective, the biggest problem is that tissue samples for TIL production cannot be obtained from all cancer patients. Even worse, in some cases, TILs cannot be isolated from resected tumor tissues. Recently, with the help of lentivirus and other gene engineering technologies, a specific TCR recognizing tumor antigen was inserted into the genome of bulk T cells, which makes it possible to produce antigen-specific T cells for everyone. Thus, TCR-engineered T cells (TCR-Ts) are therefore considered as a more promising treatment for cancer patients.
Lots of TCR-Ts are currently tested in clinical trials as reviewed by other groups (20-22). In 2006, Morgan et al. is the first group to report clinical trial results to demonstrate the feasibility of treating tumors with TCR-Ts (23). In that study two patients showed a sustained objective regression of their metastatic melanoma and remained clinically disease-free for at least 20 months. In 2009, Johnson et al. used TCR-Ts to target MART-1 (melanoma antigen recognized by T cells 1) and gp100 for the treatment of melanoma (24). The response rate of targeting MART-1 was 30% (6/20), and the response rate of targeting gp100 was 19% (3/16). Moreover, in 2015, Robbins et al. reported that the response rates of NY-ESO-1 (New York esophageal squamous cell carcinoma 1)-specific TCR-engineered autologous T cells against synovial sarcoma and melanoma was 61% (11/18) and 55% (11/20), respectively (25,26). Nowadays, more clinal trials of TCR-Ts are ongoing. The majority of the TCR-T targets used in these trials are NY-ESO-1, MAGE family (melanoma antigen family), AFP (alpha fetoprotein), WT-1 (Wilms’ tumor antigen 1), HPV (human papilloma virus), etc.
Expanding T cell epitope reservoir for CTLs-based cancer immunotherapy
The peptide-HLA complexes recognized by TCR are called T cell epitopes. Known T cell epitopes targeted by TCR-Ts which have been tested in clinical trials were summarized in Table 1 of this review. Up to date, as shown in the list, a very limited number of T cell epitopes have been identified and targeted in immunotherapy. Most of known peptides are restricted by HLA*02:01. Therefore, identifying new targets for CTLs-based immunotherapy, especially peptides restricted by other HLA, is urgently needed in order to further enhance the therapeutic value of TCR-Ts. Although T cell epitopes can be identified by sequencing, bioinformatics and mass spectrometry, identifying new antigens that are immunogenic and can promote tumor rejection remains a major challenge (49).
Table 1
| Antigen | HLA | Sequence | Cancer (s) | Reference number |
|---|---|---|---|---|
| p53 | HLA-A*02:01 | LLGRNSFEV | Metastatic melanoma | ( |
| MART1 | HLA-A*02:01 | AAGIGILTV | Metastatic melanoma | ( |
| MART1 | HLA-A*02:01 | EAAGIGILTV | Stage IV skin melanoma, eye melanoma | ( |
| gp100 | HLA-A*02:01 | KTWGQYWQV | Metastatic melanoma | ( |
| NY-ESO-1 | HLA-A*02:01, HLA-A*02:05, HLA-A*02:06 | SLLMWITQC | Metastatic melanoma, Metastatic SCS, solid cancers | ( |
| CEA | HLA-A*02:01 | IMIGVLVGV | Metastatic CRC | ( |
| MAGE-A3 | HLA-A*02:01 | KVAELVHFL | Melanoma, SCS; breast, cervical, renal, bladder cancers | ( |
| MAGE-A3 | HLA-A*01 | EVDPIGHLY | High-risk or relapsed myeloma | ( |
| MAGE-A4 | HLA-A*24:02 | NYKRCFPVI | Solid cancers | ( |
| MAGE-A4 | HLA-A*02 | GVYDGREHTV | Solid and hematological malignancies | ( |
| MAGE-A10 | HLA-A*02:01, HLA-A*02:06 | GLYDGMEHL | Advanced NSCLC | ( |
| MAGE-A12 | HLA-A*02:01 | KMVELVHFL | Esophageal cancer | ( |
| WT1 | HLA-A*02:01 | RMFPNAPYL | MDS, AML | ( |
| Tyrosinase | HLA-A*02:01 | YMDGTMSQV, YMNGTMSQV | Melanoma | ( |
| HPV E6 | HLA-A*02:01 | TIHDIILECV | HPV-associated cancers | ( |
| HPV E7 | HLA-A*02:01 | YMLDLQPET | HPV-associated cancers | ( |
| Human thyroglobulin(hTG) | HLA-A*02:01 | SKYISSLKTSADG | Metastatic thyroid cancer | ( |
| PRAME | HLA-A*02:01, HLA-A2*02:01 | VLDGLDVLL, SLYSFPEPEA, ALYVDSLFFL, SLLQHLIGL | AML, MDS, uveal melanoma | ( |
| KRAS G12V | HLA-A*11:01 | VVGAVGVGK | Pancreatic, gastric, gastrointestinal, colon, rectal cancers | ( |
| KRAS G12D | HLA-A*11:01 | VVGADGVGK | Pancreatic, gastric, gastrointestinal, colon, rectal cancers | ( |
| HA-1 | HLA-A*02:01 | VLHDDLLEA | Relapsed or refractory acute Leukemia | ( |
| TGFβRII frameshift protein | HLA-A*02 | RLSSCVPVA | CRC | ( |
| AFP | HLA-A*02:01, HLA-A*02:642 | FMNKFIYEI | HCC | ( |
AFP, alpha-fetoprotein; CEA, carcinoembryonic antigen; HA-1, minor histocompatibility (H) antigen; HPV, human papilloma virus; HERV-E-derived antigen, human endogenous retrovirus-derived antigen; MART-1, melanoma antigen recognized by T cells 1; NY-ESO-1, New York esophageal squamous cell carcinoma 1; PRAME, preferentially expressed antigen in melanoma; TGFβRII, transforming growth factor beta receptor type II; WT1, Wilms’ tumor antigen; AML, acute myeloid leukemia; ccRCC, clear cell renal cell carcinoma; CRC, colorectal cancer; HCC, hepatocellular cancer; MDS, myelodysplastic syndrome; SCS, synovial cell sarcoma.
Shared antigens expressed by broad types of cancers or individuals are considered limited. Therefore, targeting unique, patient-specific tumor mutations now has attracted a lot of attention and could become a promising alternative in the near future. Moreover, targeting “neoantigens”, the somatic mutations expressed only by tumor cells, might enable specific tumor destruction without causing off-target damage to vital healthy tissues (50). Neoantigen-reactive T cells have been administered to cancer patients and shown objective response. In some cases, complete regressions in patients with different types of cancers were also observed (51-53). In order to identify neoantigens, tumor sections and corresponding normal tissues were sent for whole-exome sequencing to determine the tumor-specific non-synonymous mutations in protein-coding regions. After that, candidate neoantigens are selected according to HLA-binding affinity, expression level, variant allele frequency and several other criteria. Finally, the immunogenicity of the selected candidate peptides is evaluated with different immunological screening assays (54).
Next generation sequencing has greatly facilitated the progress of TCR-based immunotherapy. A large number of candidates neoepitopes could be identified through whole-exome sequencing. Moreover, TCR clones could also be discovered by high throughput single cell sequencing (55,56). The traditional immunological screening assays for T cell antigen discovery, such as interferon-γ (IFN-γ) ELISA/ELISPOT and pHLA multimer staining, are usually laborious and time-consuming. Recently several high throughput techniques have been developed to identify cognate antigens for T cells (57-64). The research team led by Dr. Stephen J. Elledge has developed a high throughput, whole-genome screening platform called T-Scan to identify antigens recognized by T cells (60). In this study, this technology was applied to identify multiple cytomegalovirus (CMV) antigens that can be recognized by memory T cells. Moreover, they also used this technology to successfully discover the genome-wide targets of self-reactive TCRs. David Baltimore’s research team developed two cell-based platforms for TCR antigen discovery. One platform used chimeric receptors called signaling and antigen-presenting bifunctional receptors (SABRs) (58). These chimeric receptors are composed of an extracellular pHLA fused to an intracellular CD3ζ signaling domain and a CD28 co-stimulatory domain. When recognized by a specific TCR, this interaction triggers the expression of green fluorescent protein (GFP) and CD69 on Nuclear Factor of Activated T cells (NFAT)-GFP-Jurkat cells which can be selected and sequenced to identify the specific peptide recognized. The other platform exploits a membrane transfer phenomenon called trogocytosis, a rapid exchange of envelope fragments or related molecules between cells through cell-to-cell contact (59). Co-incubation of T cells expressing an orphan TCR with target cells led to specific labeling of cognate target cells, enabling isolation of these target cells and sequencing of the cognate TCR ligand. These high throughput techniques not only facilitate the screening of tumor antigen targets, but also can be used to discover the off-target reactivities of a therapeutic candidate TCR, making it a versatile tool for the development of T cell immunotherapy.
Tumor may escape CTLs-based immunotherapy through downregulation of HLA
Tumor immune escape refers to the phenomenon that in order to survive and proliferate in human bodies, tumor cells can escape from the surveillance of immune system. The downregulation or loss of HLA class I molecules is an important mechanism for tumors to escape from T cell-mediated immune responses.
HLA expression changes are a common event in the carcinogenesis process, and generally occur at the early stage (65,66). The downregulation or loss of HLA class I molecules can prevent tumor cells from being recognized by CTL. Besides, it was reported that class I molecules can be used as tumor suppressor genes in melanoma. Down-regulating the gene enhanced the carcinogenicity of cells, and allow melanoma cells to have a higher proliferation rate and greater migration and invasion potential (67). There are mainly two types of HLA downregulation. The first one is total HLA class I loss or downregulation, which may due to the mutation of beta-2 microglobulin (b2m) gene, complete loss of HLA class I locus or defects in antigen processing and transport pathway. The other one is partial HLA class I loss or downregulation, due to loss of one or several HLA alleles or epigenetically downregulated HLA gene expression. HLA downregulation is common in cancers (68). The percentage of total or partial HLA loss ranges from 65% to 90%, depending on the type of cancer (69). Carretero et al. examined the expression of HLA class I antigens in ten metastatic lesions obtained from a melanoma patient undergoing immunotherapy. The eight regressing metastases showed high level of HLA class I expression, whereas the two progressing lesions had low levels (70). Therefore, HLA class I downregulation may be barriers for effective CTLs-based immunotherapy.
Immunotherapies to overcome HLA downregulation
Chimeric antigen receptor (CAR) T-cell (CAR-T)
The discovery of CAR-T therapy provided a way to get around the limitation of the dependence on class I molecules. The main concept is to trigger T-lymphocyte cytotoxic reaction without the need for HLA recognition. CAR is a genetically engineered hybrid of an antibody and a TCR (71). CAR confers the T cells the abilities to recognize tumor cells with a chosen surface antigen and to trigger T cell activation with TCR signaling pathway.
The CAR structure consists of three parts, the extracellular antigen-binding region, the intracellular signal peptide region, and the transmembrane region. Its extracellular antigen-binding region is composed of single-chain variable fragments (scFv) derived from an antibody; the transmembrane region connects intracellular and extracellular structures, usually comprising CD8 or IgG4-Fc; and the intracellular signal peptide region, usually carrying a co-stimulatory domain and a CD3ζ chain, is mainly responsible for T cell activation. The CAR structure has gone through four generations of development so far. In the first-generation CAR, a scFv was fused to the gamma chain of an immunoglobulin or the zeta chain of a CD3 complex. There was no costimulatory molecule, and the survival time in vivo was short (72,73). The second generation was improved one step further by fusing a costimulatory molecule to the upstream region of a CD3 domain, mainly CD28 or 4-1BB (74,75). The third generation CAR combines multiple co-stimulatory domains, such as CD28 and 4-1BB or CD28 and OX40. The fourth-generation CAR-T further adds factors that enhance T cell expansion, persistence, and anti-tumoral activity, such as IL-2, IL-5, IL-12 and co-stimulatory ligands (76).
Immunotherapy with CAR-T cells has achieved tremendous successes in treatment of hematological malignancies. By targeting CD19, two second-generation CAR-Ts were approved by the US Food and Drug Administration (FDA) for treating leukemia and lymphoma in 2017, of which Kymriah uses 4-1BB as a co-stimulation domain and Yescarta uses CD28 as a co-stimulation domain. Researchers found that patients treated with tisagenlecleucel (Kymriah®) for relapsed/refractory B-cell precursor acute lymphoblastic leukemia (r/r ALL) showed a response rate that exceeds the response rate reported previously for standard chemotherapies. Moreover, clinical data showed that tisagenlecleucel has a continuous response without major safety concern (71,77).
There are also several ongoing clinical investigations about CAR-T therapy in solid tumors. The popular targets include glypican-3 (GPC3) (78,79), ganglioside GD2 (80,81), EGFR (82,83), EGFRvIII (84,85), etc. However, developing CAR-Ts for the treatment of solid tumors is challenging. Compared to the success in hematological malignancies, CAR-T therapy has to date been much less effective for solid tumors (86,87). The first obstacle in treating solid tumors with CAR-T cells is that there are limited antigens solely expressed on the cell surface of tumor cells but not normal cells. In the treatment of neuroblastoma, the fatal neurotoxicity was observed in high-affinity GD2-specific CAR-T cell therapy because of low amounts of GD2 expression in the cerebellum and basal regions of the brain (88). These results highlight the challenges associated with target antigens that exhibit shared expression on critical normal tissues. Therefore, to improve target specificity and eliminate toxicity are necessary in CAR-T cell therapy. Another problem for CAR-T therapy is the poor infiltration and survival of CAR-T cells in tumor microenvironment. To solve these problems, Ma et al. enhanced CAR-T cell activity against solid tumors by vaccine boosting. They used a CAR targeting EGFRvIII in combination with a vaccine containing an amphiphilic polymer linked to the EGFRvIII target antigen. In mice with EGFRvIII+ gliomas, vaccination resulted in improved CAR-T cell proliferation and survival, and improved infiltration of activated CAR-T cells into tumor sites (89,90).
Natural killer cells (NK cells)
NK cells are another type of cytotoxic lymphocytes, whose function is also mediated by the interaction of cell surface receptors with HLA class I molecules (91).
NK cells circulate in peripheral blood and are larger than T cells in size, and their phenotypes are different from that of T cells. There are no immunoglobulins or TCRs on the surface of NK cells. Human NK cells are generally defined as cells that lack the cell surface marker CD3 and express CD16 and/or CD56 cell surface glycoproteins. Immediately after pathogen infection, NK cells migrate to the site of inflammation and release their immune function. In addition to cytotoxic functions, NK cells can also secrete certain cytokines to kill infected cells.
NK cell-mediated cell killing activity was inversely related to the expression of HLA class I molecules. The mechanism is that when the specific receptor of the HLA class I molecule on the surface of NK cells encounters its ligand molecule, it will send an inhibitory signal to prevent NK cells from being activated to exert cytotoxicity and secret cytokines (92), preventing NK cells from killing healthy cells. To that end, each NK cell expresses at least one inhibitory receptor that can specifically interact with type I molecules. There are two types of inhibitory receptors in human NK cells for HLA class I molecules, one is a heterodimer composed of membrane molecules CD94 and NKG2A that are covalently bound through disulfide bonds (93,94). The other type is called killer-cell immunoglobulin-like receptor (KIR) which are a family of transmembrane glycoproteins expressed on NK cells and a subset of T cells. The different ways used by NK cells and CD8+ T cells to recognize and respond to class I molecules allow them to collectively generate complementary immune defense against infection.
Since 1970s, several studies have revealed the important role played by NK cells in anti-tumor cytotoxicity. For examples, NK cells can release CCL5, XCL1, and XCL2 to promote the aggregation of DCs inside solid tumors and to promote the antitumor effect of CD8+ T cells (95). In a recent study, scientists transformed NK cells derived from patients with ovarian cancer into a cytotoxic CD56superbrightCD16+ subset, which can effectively control the growth of autologous ovarian cancer xenografts in mice (96).
NK cells function through an antigen-independent pathway and can recognize the loss of HLA molecules as an activation signal, which effectively reduces the possibility of immune escape of tumor cells due to the downregulation of class I molecules. By exploiting this feature, we can then utilize NK cells to make up for the restriction on target recognition imposed by HLA class I molecules in TCR-T therapy.
CD4+ T cells
At present, most TCR-T researches are focused on CD8+ T cells. Since tumor cells often escape the surveillance of the immune system by down-regulating the expression of class I molecules, the application of CD8+-associated TCR-T therapy is greatly limited. Therefore, some researchers have turned to CD4+ T cells.
Class II molecules mainly present exogenous antigens and are only selectively expressed on specialized APCs that are functionally differentiated, such as macrophages, B cells, and DCs. One common feature shared by these cells is that class II molecules and antigen peptides form complexes before being transferred to the cell membrane surface and subsequently recognized and bound by CD4+ T cells.
CD4+ T cells are also called T helper cells, which help the activation of CTLs, B cells, macrophages and DCs. CD4+ T cells can differentiate into different subsets such as Th1, Th2, Th17 and Treg cells. Although CD8+ CTLs are the preferred tools to target tumors, CD4+ T cells are also required for effective antitumor immunity (97,98). On one hand, CD4+ T cells could kill tumor cells directly when HLA class II molecules expression were induced on certain tumor cells by IFN-γ stimulation (98). On the other hand, CD4+ T cells can exert indirect cytotoxicity. Tumor antigen was processed by HLA class II positive APCs and presented to CD4+ T cells. Tumor-specific CD4+ T cells were activated and started to secrete cytokines (99,100). Vaccination with CD4+ immunogenic mutations/neo-epitope induced cytotoxic T lymphocyte responses and conferred antitumor activity both in mice and patients, which revealed the participation of CD4+ T cells in immunotherapy (100,101). Moreover, Robert D. Schreiber’s group found that CD4+ T cells were also required in immune checkpoint therapy and the expression of MHC class II-restricted antigens by tumor cells was required at the site of successful rejection, indicating that activation of CD4+ T cells must also occur in the tumor microenvironment (102).
Up to date, several studies have shown that adoptive transfer of CD4+ T cells can also induce tumor regression (103-105). Researchers have generated an MHC class II-restricted TCR transgenic mouse model in which CD4+ T cells recognize one epitope in tyrosinase-related protein 1 (TRP-1), an antigen expressed by normal melanocytes and B16 murine melanoma (103). Both in vitro and in vivo experiments have confirmed that CD4+ T cells can eliminate established B16 melanoma, and its therapeutic effect is mainly mediated by IFN-γ. In a clinical study (106) patients with metastatic melanoma were treated with autologous DP4-restricted NY-ESO-1 specific CD4+ T cell clones and have achieved a long-term complete response for over 2 years, suggesting that CD4+ T cells can also induce long-term tumor regression in human similar to what CD8+ T cells have achieved.
Frequently found in a variety of cancer types, MAGE-A3 is a cancer germline antigen and is one of the best targets for cancer immunotherapy. In a clinical study conducted in 2017, 17 patients received adoptive transfer of CD4+ T cells retrovirally transduced with MAGE-A3 TCR after lymphadenectomy plus systemic high-dose IL-2. The results showed that objective complete remission was observed in patients with metastatic cervical cancer. Patients with esophageal cancer, urothelial cancer, and osteosarcoma all had objective responses with durations ≥4 months. There were no treatment-associated adverse effects. Taken together, these findings have proven the safety and effectiveness of the MAGE-A3 specific CD4+ TCR-Ts (107).
Summary
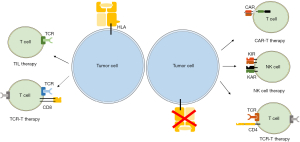
HLA molecules play a pivotal role in T cell-mediated adaptive immunity. HLA class I molecules exist on most types of human cells and interact with TCRs to activate T cells to induce adaptive immune responses. CD8+ T cell-based therapies such as TIL therapy and TCR-T therapy are HLA-dependent immunotherapies (Figure 2). Although TIL has achieved significant results in the treatment of metastatic melanoma, it is still difficult to isolate and identify effective TILs for other malignant tumors. In addition, as a “personalized” treatment, its industrial manufacturing process is full of obstacles. The current researches on TIL therapy are focused on how to use a rapid method to isolate, identify, and expand TILs to the clinically required doses. In TCR-T therapy, antigen-specific TCRs are transduced into normal T cells using retrovirus or lentivirus making it more convenient compared to TILs. Currently, several TCR-Ts are under investigation in clinical trials to further explore the therapeutic value. However, tumor cells may escape T cell attacks through downregulation of HLA, which poses a greater challenge to HLA-dependent immunotherapy. Thus, HLA-independent immunotherapies such as CAR-T therapy, NK therapy and CD4+ T cell therapy are discussed above (Figure 2). Interestingly, Crowther et al. recently found one TCR recognized and killed most human cancer types via the monomorphic MHC class I-related protein, MR1 (108). This MR1-restricted TCR mediated in vivo regression of tumor both in mice and in melanoma patients without the requirement of a specific HLA. These findings offered another opportunity for HLA-independent and pan-population immunotherapies. Considering great heterogeneity in tumor microenvironment, we believe that the combination of different immune cell therapies such as CD8+ T cells and CD4+ T cells should be a trend of tumor adoptive immunotherapy in the future.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (81903156).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aob-20-27). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kruger S, Ilmer M, Kobold S, et al. Advances in cancer immunotherapy 2019 - latest trends. J Exp Clin Cancer Res 2019;38:268. [Crossref] [PubMed]
- Jensen PE. Mechanisms of antigen presentation. Clin Chem Lab Med 1999;37:179-86. [Crossref] [PubMed]
- Parham P. Virtual reality in the MHC. Immunol Rev 1999;167:5-15. [Crossref] [PubMed]
- Thorsby E. A short history of HLA. Tissue Antigens 2009;74:101-16. [Crossref] [PubMed]
- Carey BS, Poulton KV, Poles A. Factors affecting HLA expression: A review. Int J Immunogenet 2019;46:307-20. [Crossref] [PubMed]
- Robinson J, Halliwell JA, McWilliam H, et al. IPD--the Immuno Polymorphism Database. Nucleic Acids Res 2013;41:D1234-40. [Crossref] [PubMed]
- Chowell D, Krishna C, Pierini F, et al. Evolutionary divergence of HLA class I genotype impacts efficacy of cancer immunotherapy. Nat Med 2019;25:1715-20. [Crossref] [PubMed]
- Bouvier M, Wiley DC. Importance of peptide amino and carboxyl termini to the stability of MHC class I molecules. Science 1994;265:398-402. [Crossref] [PubMed]
- Wieczorek M, Abualrous ET, Sticht J, et al. Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front Immunol 2017;8:292. [Crossref] [PubMed]
- Boyle LH, Hermann C, Boname JM, et al. Tapasin-related protein TAPBPR is an additional component of the MHC class I presentation pathway. Proc Natl Acad Sci U S A 2013;110:3465-70. [Crossref] [PubMed]
- Hermann C, van Hateren A, Trautwein N, et al. TAPBPR alters MHC class I peptide presentation by functioning as a peptide exchange catalyst. Elife 2015;4:e09617 [Crossref] [PubMed]
- Chicz RM, Urban RG, Lane WS, et al. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature 1992;358:764-8. [Crossref] [PubMed]
- Yamashita Y, Anczurowski M, Nakatsugawa M, et al. HLA-DP(84Gly) constitutively presents endogenous peptides generated by the class I antigen processing pathway. Nat Commun 2017;8:15244. [Crossref] [PubMed]
- Embgenbroich M, Burgdorf S. Current Concepts of Antigen Cross-Presentation. Front Immunol 2018;9:1643. [Crossref] [PubMed]
- Joffre OP, Segura E, Savina A, et al. Cross-presentation by dendritic cells. Nat Rev Immunol 2012;12:557-69. [Crossref] [PubMed]
- Rock KL, Reits E, Neefjes J. Present Yourself! By MHC Class I and MHC Class II Molecules. Trends Immunol 2016;37:724-37. [Crossref] [PubMed]
- Maher J, Davies ET. Targeting cytotoxic T lymphocytes for cancer immunotherapy. Br J Cancer 2004;91:817-21. [Crossref] [PubMed]
- Rosenberg SA, Packard BS, Aebersold PM, et al. Use of tumor-infiltrating lymphocytes and interleukin-2 in the immunotherapy of patients with metastatic melanoma. A preliminary report. N Engl J Med 1988;319:1676-80. [Crossref] [PubMed]
- Rohaan MW, van den Berg JH, Kvistborg P, et al. Adoptive transfer of tumor-infiltrating lymphocytes in melanoma: a viable treatment option. J Immunother Cancer 2018;6:102. [Crossref] [PubMed]
- Johnson LA, June CH. Driving gene-engineered T cell immunotherapy of cancer. Cell Res 2017;27:38-58. [Crossref] [PubMed]
- Zhang J, Wang L. The Emerging World of TCR-T Cell Trials Against Cancer: A Systematic Review. Technol Cancer Res Treat 2019;18:1533033819831068 [Crossref] [PubMed]
- Zhao L, Cao YJ, Engineered T. Cell Therapy for Cancer in the Clinic. Front Immunol 2019;10:2250. [Crossref] [PubMed]
- Morgan RA, Dudley ME, Wunderlich JR, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science 2006;314:126-9. [Crossref] [PubMed]
- Johnson LA, Morgan RA, Dudley ME, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood 2009;114:535-46. [Crossref] [PubMed]
- Robbins PF, Morgan RA, Feldman SA, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol 2011;29:917-24. [Crossref] [PubMed]
- Robbins PF, Kassim SH, Tran TL, et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin Cancer Res 2015;21:1019-27. [Crossref] [PubMed]
- Antonia SJ, Mirza N, Fricke I, et al. Combination of p53 cancer vaccine with chemotherapy in patients with extensive stage small cell lung cancer. Clin Cancer Res 2006;12:878-87. [Crossref] [PubMed]
- Borbulevych OY, Santhanagopolan SM, Hossain M, et al. TCRs used in cancer gene therapy cross-react with MART-1/Melan-A tumor antigens via distinct mechanisms. J Immunol 2011;187:2453-63. [Crossref] [PubMed]
- Gannon PO, Baumgaertner P, Huber A, et al. Rapid and Continued T-Cell Differentiation into Long-term Effector and Memory Stem Cells in Vaccinated Melanoma Patients. Clin Cancer Res 2017;23:3285-96. [Crossref] [PubMed]
- Smith FO, Downey SG, Klapper JA, et al. Treatment of metastatic melanoma using interleukin-2 alone or in conjunction with vaccines. Clin Cancer Res 2008;14:5610-8. [Crossref] [PubMed]
- D'Angelo SP, Melchiori L, Merchant MS, et al. Antitumor Activity Associated with Prolonged Persistence of Adoptively Transferred NY-ESO-1 (c259)T Cells in Synovial Sarcoma. Cancer Discov 2018;8:944-57. [Crossref] [PubMed]
- Rosati SF, Parkhurst MR, Hong Y, et al. A novel murine T-cell receptor targeting NY-ESO-1. J Immunother 2014;37:135-46. [Crossref] [PubMed]
- Parkhurst MR, Joo J, Riley JP, et al. Characterization of genetically modified T-cell receptors that recognize the CEA:691-699 peptide in the context of HLA-A2.1 on human colorectal cancer cells. Clin Cancer Res 2009;15:169-80. [Crossref] [PubMed]
- Chinnasamy N, Wargo JA, Yu Z, et al. A TCR targeting the HLA-A*0201-restricted epitope of MAGE-A3 recognizes multiple epitopes of the MAGE-A antigen superfamily in several types of cancer. J Immunol 2011;186:685-96. [Crossref] [PubMed]
- Cameron BJ, Gerry AB, Dukes J, et al. Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells. Sci Transl Med 2013;5:197ra103 [Crossref] [PubMed]
- Miyahara Y, Naota H, Wang L, et al. Determination of cellularly processed HLA-A2402-restricted novel CTL epitopes derived from two cancer germ line genes, MAGE-A4 and SAGE. Clin Cancer Res 2005;11:5581-9. [Crossref] [PubMed]
- Kageyama S, Ikeda H, Miyahara Y, et al. Adoptive Transfer of MAGE-A4 T-cell Receptor Gene-Transduced Lymphocytes in Patients with Recurrent Esophageal Cancer. Clin Cancer Res 2015;21:2268-77. [Crossref] [PubMed]
- Border EC, Sanderson JP, Weissensteiner T, et al. Affinity-enhanced T-cell receptors for adoptive T-cell therapy targeting MAGE-A10: strategy for selection of an optimal candidate. Oncoimmunology 2018;8:e1532759 [Crossref] [PubMed]
- Thomas S, Xue SA, Cesco-Gaspere M, et al. Targeting the Wilms tumor antigen 1 by TCR gene transfer: TCR variants improve tetramer binding but not the function of gene modified human T cells. J Immunol 2007;179:5803-10. [Crossref] [PubMed]
- Moore T, Wagner CR, Scurti GM, et al. Correction to: Clinical and immunologic evaluation of three metastatic melanoma patients treated with autologous melanoma-reactive TCR-transduced T cells. Cancer Immunol Immunother 2018;67:327. [Crossref] [PubMed]
- Draper LM, Kwong ML, Gros A, et al. Targeting of HPV-16+ Epithelial Cancer Cells by TCR Gene Engineered T Cells Directed against E6. Clin Cancer Res 2015;21:4431-9. [Crossref] [PubMed]
- Jin BY, Campbell TE, Draper LM, et al. Engineered T cells targeting E7 mediate regression of human papillomavirus cancers in a murine model. JCI Insight 2018;3:e99488 [Crossref] [PubMed]
- Sánchez A, Cardona R, Munera M, et al. Identification of antigenic epitopes of thyroperoxidase, thyroglobulin and interleukin-24. Exploration of cross-reactivity with environmental allergens and possible role in urticaria and hypothyroidism. Immunol Lett 2020;220:71-8. [Crossref] [PubMed]
- Amir AL, van der Steen DM, van Loenen MM, et al. PRAME-specific Allo-HLA-restricted T cells with potent antitumor reactivity useful for therapeutic T-cell receptor gene transfer. Clin Cancer Res 2011;17:5615-25. [Crossref] [PubMed]
- Wang QJ, Yu Z, Griffith K, et al. Identification of T-cell Receptors Targeting KRAS-Mutated Human Tumors. Cancer Immunol Res 2016;4:204-14. [Crossref] [PubMed]
- Dossa RG, Cunningham T, Sommermeyer D, et al. Development of T-cell immunotherapy for hematopoietic stem cell transplantation recipients at risk of leukemia relapse. Blood 2018;131:108-20. [Crossref] [PubMed]
- Inderberg EM, Walchli S, Myhre MR, et al. T cell therapy targeting a public neoantigen in microsatellite instable colon cancer reduces in vivo tumor growth. Oncoimmunology 2017;6:e1302631 [Crossref] [PubMed]
- Docta RY, Ferronha T, Sanderson JP, et al. Tuning T-Cell Receptor Affinity to Optimize Clinical Risk-Benefit When Targeting Alpha-Fetoprotein-Positive Liver Cancer. Hepatology 2019;69:2061-75. [Crossref] [PubMed]
- Abelin JG, Keskin DB, Sarkizova S, et al. Mass Spectrometry Profiling of HLA-Associated Peptidomes in Mono-allelic Cells Enables More Accurate Epitope Prediction. Immunity 2017;46:315-26. [Crossref] [PubMed]
- Yamamoto TN, Kishton RJ, Restifo NP. Developing neoantigen-targeted T cell-based treatments for solid tumors. Nat Med 2019;25:1488-99. [Crossref] [PubMed]
- Zacharakis N, Chinnasamy H, Black M, et al. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat Med 2018;24:724-30. [Crossref] [PubMed]
- Tran E, Robbins PF, Lu YC, et al. T-Cell Transfer Therapy Targeting Mutant KRAS in Cancer. N Engl J Med 2016;375:2255-62. [Crossref] [PubMed]
- Strønen E, Toebes M, Kelderman S, et al. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science 2016;352:1337-41. [Crossref] [PubMed]
- Garcia-Garijo A, Fajardo CA, Gros A. Determinants for Neoantigen Identification. Front Immunol 2019;10:1392. [Crossref] [PubMed]
- De Simone M, Rossetti G, Pagani M. Single Cell T Cell Receptor Sequencing: Techniques and Future Challenges. Front Immunol 2018;9:1638. [Crossref] [PubMed]
- Singh M, Al-Eryani G, Carswell S, et al. High-throughput targeted long-read single cell sequencing reveals the clonal and transcriptional landscape of lymphocytes. Nat Commun 2019;10:3120. [Crossref] [PubMed]
- Gee MH, Han A, Lofgren SM, et al. Antigen Identification for Orphan T Cell Receptors Expressed on Tumor-Infiltrating Lymphocytes. Cell 2018;172:549-563.e16. [Crossref] [PubMed]
- Joglekar AV, Leonard MT, Jeppson JD, et al. T cell antigen discovery via signaling and antigen-presenting bifunctional receptors. Nat Methods 2019;16:191-8. [Crossref] [PubMed]
- Li G, Bethune MT, Wong S, et al. T cell antigen discovery via trogocytosis. Nat Methods 2019;16:183-90. [Crossref] [PubMed]
- Kula T, Dezfulian MH, Wang CI, et al. T-Scan: A Genome-wide Method for the Systematic Discovery of T Cell Epitopes. Cell 2019;178:1016-1028.e13. [Crossref] [PubMed]
- Lanzarotti E, Marcatili P, Nielsen M. T-Cell Receptor Cognate Target Prediction Based on Paired alpha and beta Chain Sequence and Structural CDR Loop Similarities. Front Immunol 2019;10:2080. [Crossref] [PubMed]
- Lu YC, Zheng Z, Robbins PF, et al. An Efficient Single-Cell RNA-Seq Approach to Identify Neoantigen-Specific T Cell Receptors. Mol Ther 2018;26:379-89. [Crossref] [PubMed]
- Riley TP, Keller GLJ, Smith AR, et al. Structure Based Prediction of Neoantigen Immunogenicity. Front Immunol 2019;10:2047. [Crossref] [PubMed]
- Sharma G, Rive CM, Holt RA. Rapid selection and identification of functional CD8(+) T cell epitopes from large peptide-coding libraries. Nat Commun 2019;10:4553. [Crossref] [PubMed]
- Campoli M, Ferrone S. HLA antigen changes in malignant cells: epigenetic mechanisms and biologic significance. Oncogene 2008;27:5869-85. [Crossref] [PubMed]
- Mendez R, Aptsiauri N, Del Campo A, et al. HLA and melanoma: multiple alterations in HLA class I and II expression in human melanoma cell lines from ESTDAB cell bank. Cancer Immunol Immunother 2009;58:1507-15. [Crossref] [PubMed]
- Garrido C, Paco L, Romero I, et al. MHC class I molecules act as tumor suppressor genes regulating the cell cycle gene expression, invasion and intrinsic tumorigenicity of melanoma cells. Carcinogenesis 2012;33:687-93. [Crossref] [PubMed]
- Bubeník J. Tumour MHC class I downregulation and immunotherapy Oncol Rep 2003;10:2005-8. (Review). [PubMed]
- Garrido F, Aptsiauri N, Doorduijn EM, et al. The urgent need to recover MHC class I in cancers for effective immunotherapy. Curr Opin Immunol 2016;39:44-51. [Crossref] [PubMed]
- Carretero R, Romero JM, Ruiz-Cabello F, et al. Analysis of HLA class I expression in progressing and regressing metastatic melanoma lesions after immunotherapy. Immunogenetics 2008;60:439-47. [Crossref] [PubMed]
- Subklewe M, von Bergwelt-Baildon M, Humpe A. Chimeric Antigen Receptor T Cells: A Race to Revolutionize Cancer Therapy. Transfus Med Hemother 2019;46:15-24. [Crossref] [PubMed]
- Brocker T, Karjalainen K. Signals through T cell receptor-zeta chain alone are insufficient to prime resting T lymphocytes. J Exp Med 1995;181:1653-9. [Crossref] [PubMed]
- Gong MC, Latouche JB, Krause A, et al. Cancer patient T cells genetically targeted to prostate-specific membrane antigen specifically lyse prostate cancer cells and release cytokines in response to prostate-specific membrane antigen. Neoplasia 1999;1:123-7. [Crossref] [PubMed]
- Krause A, Guo HF, Latouche JB, et al. Antigen-dependent CD28 signaling selectively enhances survival and proliferation in genetically modified activated human primary T lymphocytes. J Exp Med 1998;188:619-26. [Crossref] [PubMed]
- Porter DL, Levine BL, Kalos M, et al. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med 2011;365:725-33. [Crossref] [PubMed]
- Chmielewski M, Abken H. TRUCKs: the fourth generation of CARs. Expert Opin Biol Ther 2015;15:1145-54. [Crossref] [PubMed]
- Forsberg MH, Das A, Saha K, et al. The potential of CAR T therapy for relapsed or refractory pediatric and young adult B-cell ALL. Ther Clin Risk Manag 2018;14:1573-84. [Crossref] [PubMed]
- Batra SA, Rathi P, Guo L, et al. Glypican-3-specific CAR T cells co-expressing IL15 and IL21 have superior expansion and antitumor activity against hepatocellular carcinoma. Cancer Immunol Res 2020;8:309-20. [Crossref] [PubMed]
- Jiang Z, Jiang X, Chen S, et al. Anti-GPC3-CAR T Cells Suppress the Growth of Tumor Cells in Patient-Derived Xenografts of Hepatocellular Carcinoma. Front Immunol 2017;7:690. [Crossref] [PubMed]
- Yu J, Wu X, Yan J, et al. Anti-GD2/4-1BB chimeric antigen receptor T cell therapy for the treatment of Chinese melanoma patients. J Hematol Oncol 2018;11:1. [Crossref] [PubMed]
- Gargett T, Yu W, Dotti G, et al. GD2-specific CAR T Cells Undergo Potent Activation and Deletion Following Antigen Encounter but can be Protected From Activation-induced Cell Death by PD-1 Blockade. Mol Ther 2016;24:1135-49. [Crossref] [PubMed]
- Li H, Huang Y, Jiang DQ, et al. Antitumor activity of EGFR-specific CAR T cells against non-small-cell lung cancer cells in vitro and in mice. Cell Death Dis 2018;9:177. [Crossref] [PubMed]
- Ravanpay AC, Gust J, Johnson AJ, et al. EGFR806-CAR T cells selectively target a tumor-restricted EGFR epitope in glioblastoma. Oncotarget 2019;10:7080-95. [Crossref] [PubMed]
- Sahin A, Sanchez C, Bullain S, et al. Development of third generation anti-EGFRvIII chimeric T cells and EGFRvIII-expressing artificial antigen presenting cells for adoptive cell therapy for glioma. PLoS One 2018;13:e0199414 [Crossref] [PubMed]
- O'Rourke DM, Nasrallah MP, Desai A, et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci Transl Med 2017;9:eaaa0984.
- Martinez M, Moon EK. CAR T Cells for Solid Tumors: New Strategies for Finding, Infiltrating, and Surviving in the Tumor Microenvironment. Front Immunol 2019;10:128. [Crossref] [PubMed]
- Ma S, Li X, Wang X, et al. Current Progress in CAR-T Cell Therapy for Solid Tumors. Int J Biol Sci 2019;15:2548-60. [Crossref] [PubMed]
- Richman SA, Nunez-Cruz S, Moghimi B, et al. High-Affinity GD2-Specific CAR T Cells Induce Fatal Encephalitis in a Preclinical Neuroblastoma Model. Cancer Immunol Res 2018;6:36-46. [Crossref] [PubMed]
- Ma L, Dichwalkar T, Chang JYH, et al. Enhanced CAR-T cell activity against solid tumors by vaccine boosting through the chimeric receptor. Science 2019;365:162-8. [PubMed]
- Singh N, June CH. Boosting engineered T cells. Science 2019;365:119-20. [PubMed]
- Fehling HJ, Gilfillan S, Ceredig R. Alpha beta/gamma delta lineage commitment in the thymus of normal and genetically manipulated mice. Adv Immunol 1999;71:1-76. [PubMed]
- Lanier LL. NK cell receptors. Annu Rev Immunol 1998;16:359-93. [Crossref] [PubMed]
- Björkström NK, Riese P, Heuts F, et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood 2010;116:3853-64. [Crossref] [PubMed]
- Caligiuri MA. Human natural killer cells. Blood 2008;112:461-9. [Crossref] [PubMed]
- Böttcher JP, Bonavita E, Chakravarty P, et al. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell 2018;172:1022-1037.e14. [Crossref] [PubMed]
- Poznanski SM, Nham T, Chew MV, et al. Expanded CD56(superbright)CD16(+) NK Cells from Ovarian Cancer Patients Are Cytotoxic against Autologous Tumor in a Patient-Derived Xenograft Murine Model. Cancer Immunol Res 2018;6:1174-85. [Crossref] [PubMed]
- Borst J, Ahrends T, Bąbała N, et al. CD4+ T cell help in cancer immunology and immunotherapy. Nat Rev Immunol 2018;18:635-47. [Crossref] [PubMed]
- Haabeth OA, Tveita AA, Fauskanger M, et al. How Do CD4(+) T Cells Detect and Eliminate Tumor Cells That Either Lack or Express MHC Class II Molecules? Front Immunol 2014;5:174. [Crossref] [PubMed]
- Corthay A, Skovseth DK, Lundin KU, et al. Primary antitumor immune response mediated by CD4+ T cells. Immunity 2005;22:371-83. [Crossref] [PubMed]
- Kreiter S, Vormehr M, van de Roemer N, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 2015;520:692-6. [Crossref] [PubMed]
- Ott PA, Hu Z, Keskin DB, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017;547:217-21. [Crossref] [PubMed]
- Alspach E, Lussier DM, Miceli AP, et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 2019;574:696-701. [Crossref] [PubMed]
- Muranski P, Boni A, Antony PA, et al. Tumor-specific Th17-polarized cells eradicate large established melanoma. Blood 2008;112:362-73. [Crossref] [PubMed]
- Quezada SA, Simpson TR, Peggs KS, et al. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J Exp Med 2010;207:637-50. [Crossref] [PubMed]
- Hirschhorn-Cymerman D, Budhu S, Kitano S, et al. Induction of tumoricidal function in CD4+ T cells is associated with concomitant memory and terminally differentiated phenotype. J Exp Med 2012;209:2113-26. [Crossref] [PubMed]
- Hunder NN, Wallen H, Cao J, et al. Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1. N Engl J Med 2008;358:2698-703. [Crossref] [PubMed]
- Lu YC, Parker LL, Lu T, et al. Treatment of Patients With Metastatic Cancer Using a Major Histocompatibility Complex Class II-Restricted T-Cell Receptor Targeting the Cancer Germline Antigen MAGE-A3. J Clin Oncol 2017;35:3322-9. [Crossref] [PubMed]
- Crowther MD, Dolton G, Legut M, et al. Genome-wide CRISPR-Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1. Nat Immunol 2020;21:178-85. Erratum in: Nat Immunol 2020 Jun;21(6):695. [Crossref] [PubMed]
Cite this article as: Wang C, Xiong C, Hsu YC, Wang X, Chen L. Human leukocyte antigen (HLA) and cancer immunotherapy: HLA-dependent and -independent adoptive immunotherapies. Ann Blood 2020;5:14.


