
In silico molecular dynamics of human glycophorin A (GPA) extracellular structure
Introduction
Glycophorin A (GPA) is a single-pass transmembrane sialoglycoprotein found in human red blood cells (RBCs), and is one of two glycoproteins that constitute the MNS blood group system. The MNS blood group system contains multiple clinically significant antigens, and has been associated with haemolytic disease of the foetus and newborn (1).
Unfortunately, there is currently a lack of monoclonal antibodies (mAbs) available for a number of antigens in the MNS blood group system (2). Previous attempts in our laboratory at producing mAbs against various glycophorin peptides in both GPA KO mice, and biopanning experiments resulted in antibodies with strong affinity towards the peptides, but little or no affinity to an RBC. It is possible that there are structural features associated with antigenicity of GPA that are not present in the peptide sequence alone. Therefore, a structural model of the extracellular domain of GPA (GPA-ECD) may act as a stepping-stone in understanding antigen presentation, and subsequently improve the way mAbs are identified.
Typically, protein structures are solved through the process of X-ray crystallography or nuclear magnetic resonance (NMR) spectroscopy. However, the GPA-ECD is heavily glycosylated, and contains 16 O-linked and 1 N-glycosylation sites (3,4). This high level of glycosylation hinders the formation of a stable crystal for X-ray crystallography, and the high degree of heterogeneity associated with glycosylation adds further complexity when analysing any produced structures (5,6). It is possible to remove glycosylation through the use of neuraminidase cleavage or to express a soluble version of the protein in a bacterial system (7), however, there are still problems in retaining native structure through the preceding methods. NMR has been previously used to solve the transmembrane domain structure of GPA (8), however, there is limited information about the secondary structure of GPA-ECD. What is available from NMR and circular dichroism (CD) studies, which includes studies with neuraminidase cleavage of glycosylation, indicates that there is a disordered portion of GPA-ECD structure (9,10). Apart from consensus that GPA is a single-pass transmembrane protein, no detailed structure of GPA-ECD or its antigens has emerged from these methods to date.
Due to the limited availability of experimental data, combined with the difficulty of obtaining structural data for the GPA-ECD, three structure prediction methods of increasing complexity were used: (I) homology/comparative modelling, (II) threading and (III) ab initio simulation. This was performed to better understand the molecular basis of GPA antigenicity, as well as whether these in silico modelling methods can provide a prediction of GPA secondary structure and subsequent antigen conformation. Although glycosylation is a factor in antigenicity, there is limited evidence to suggest it has a significant role in protein backbone structure. Due to the increased level of complexity as well as processing power required to simulate glycosylation, this study produced an initial model of GPA-ECD structure based on an amino acid backbone. Protein glycosylation will be added once a structure has been determined in order to investigate its role in antigenicity.
Methods
Generation of 3D structures of GPA-ECD
GPA-ECD
The amino acid sequence of GPA was obtained from Uniprot (Universal Protein Resource, RRID:SCR_002380) (11) accession number P02724 (GLPA_HUMAN). The amino acid sequence of the extracellular domain of glycophorin A (GPA-ECD, residues 19–89; 71 residues) were submitted to Robetta (Robetta RRID:SCR_018805) (12), iTasser (iTasser RRID:SCR_018803) (13) and FALCON (FALCON RRID:SCR_018804) (14). The top 5–10 models generated by all three programs were used for analysis. Five ab initio models generated by Robetta (12) were further analysed and validated using atomistic molecular dynamics (MD) simulations (see “MD simulations” below). In addition, a previously predicted GPA model with a β-barrel structure (15) generated using the program FALCON (14), for the extracellular domain GPA-ECD (residues 21–91; 71 residues) was also used for MD simulations. All amino acid sequences for GYPA models can be seen in Table 1.
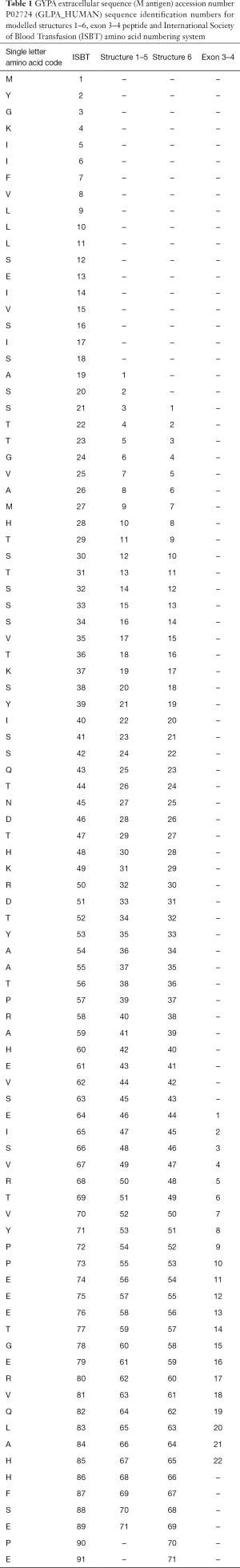
Full table
GPA-ECD exon 3–4 junction hairpin peptide
A representative structure was extracted from clustering data produced on the GPA-ECD exon 3–4 junction from the combined trajectories of the 5 Rosetta GPA-ECD models. Two structures (residues 64–85; 22 residues) were selected, in the form of a beta-hairpin with N-terminal acetyl and C-terminal amide caps added using PyMOL (PyMOL, RRID:SCR_000305) version #2.2.3 (16).
GPA-ECD exon 3–4 junction linear peptide
An extended peptide model of the GPA-ECD exon 3–4 junction (residues 64–85; 22 residues) was produced using the builder tool of PyMOL (16) with N-terminal acetyl and C-terminal amide caps added.
GPA-ECD exon 3–4 junction circular peptide
Two circular peptide models for GPA-ECD exon 3–4 junction (residues 64–85; 22 residues) were produced using Rosetta (12). The topology files were generated using a combination of GROMOS (17) and GROMACS (GROMACS, RRID:SCR_014565).
GPA-ECD exon 3–4 junction single amino acid mutations
FoldX software (FoldX, RRID:SCR_008522) (18) was used to create single amino acid changes in the GPA-ECD exon 3–4 junction hairpin peptide (produced using method above) according to known MNS antigens caused by single amino acid mutations in Table S1. The modelled structures retained the N-terminal acetyl and C-terminal amide caps.
MD simulations
All MD simulations were performed using the GPU version Gromacs 2019.1 (19) on the Wiener HPC cluster at the University of Queensland. The Gromos 54A7 force field was used to model protein structures (20,21). Each protein was placed in a cubic periodic box with the minimum distance of 1.2 nm between protein surface and wall. Each system was solvated with SPC (22) water model. The protonation states of titratable groups were chosen appropriate to pH 7.0, where N-terminal, Arg and Lys residues were protonated while C-terminal, Asp and Glu residues were deprotonated. The neutral ε-tautomer for His residues was used. No counter ions were added.
Each system (protein + water) was energy minimized and equilibrated for 200 ps with the heavy atoms of the protein positionally restrained before commencing a series of unrestrained MD simulations. All simulations were performed at constant temperature (298 K) and pressure (1 atm) using a Berendsen thermostat (coupling time of 0.1 ps) and barostat [coupling time of 1.0 ps and isothermal compressibility of 4.575×10-4 (kJ/mol/nm3)-1] (23,24). A single cut-off of 1.4 nm was used for all non-bonded interactions. The neighbour list was updated every 0.010 ps (every 5 steps). To correct for the truncation of the electrostatic interactions beyond the 1.4 nm long-range cut-off a reaction-field correction was applied using a dielectric permittivity of 78. The equations of motion were integrated using the leapfrog scheme and a step-size of 0.002 ps. Initial velocities at a given temperature were derived from a Maxwell-Boltzmann distribution. All bonds were constrained using the LINCS algorithm with a lincs_order of 4 (25). The SPC water molecules were constrained using the SETTLE algorithm (26). MD simulations were performed for 200 ns for each system (with the exception of GPA-ECD exon 3–4 junction single amino acid mutation models which were run for 100 ns); all coordinates, velocities, forces and energies were saved every 10,000 steps (20 ps) for analysis.
Analysis using RMSD, Clustering and Visual Analysis
The stability of protein structures was analysed using root mean square deviation (RMSD) as obtained by fitting the backbone atoms and calculating the RMSD for backbone atoms. Additional secondary-structure analysis was performed to check the stability and inter-change of secondary structure elements including visual analysis of the trajectory in VMD program (27) (Visual Molecular Dynamics, RRID:SCR_001820) version 1.9.3. The clustering of the relevant combined MD simulation trajectories (5 Robetta models for GPA-ECD, GPA-ECD exon 3–4 junction region as a representative hairpin, extended and circular model, and 7 FoldX GPA-ECD exon 3–4 junction mutation models), was performed using the inbuilt analysis tool in GROMACS. The samples were run for 200 ns trajectory contained 10,000 frames, with the exception of the exon 3–4 junction mutation models which were run for 100 ns for a total of 5,000 frames. The clustering method of Daura et al. [1999] as implemented in GROMACS under the name ‘GROMOS method’ was used (28). For clusters of structures in an MD simulation trajectory, the RMSD of atom positions between all pairs of structures were determined. For each structure, the number of alternate iterations either similar or dissimilar, based on backbone atom RMSD values less than or equal to a specified value determined by RMS distribution analysis, was calculated. The structure with the highest number of neighbours was taken as the centre of a cluster, and formed the complete cluster together with all its neighbours. The structures of this cluster were thereafter eliminated from the pool of structures, and the process was repeated until the pool of structures was empty. An RMS distribution plot was generated to select appropriate cut-off RMSD values in the cases where more than one unique cluster was found.
Secondary structural features were automatically identified via the internal algorithms of the Visual Molecular Dynamics program (VMD). The program feature ‘Timeline’ was used to produce a graphical representation of secondary structure associated with each amino acid across the timeline of the full MD simulation. This tool was used to determine the specific amino acids associated with secondary structures, as well as the duration of the presence of the secondary structural feature throughout the simulation.
Circular dichroism (CD) spectropolarimetry of GPA-ECD
A peptide containing the GPA extracellular domain sequence [SSTTGVAMHTSTSSSVTKSYISSQTNDTHKRDTYAATPRAHEVSEISVRTVYPPEEETGERVQLAHHFSEPE] was synthesised by Thermo Fisher. CD spectra of GPA-ECD peptide (100 µM in 20 mM KHPO4, pH 6.0) were acquired under constant N2 flush using a Jasco J-810 spectropolarimeter. Measurements were taken at 0.2-nm wavelength increments from 195 to 250 nm at 100 nm/min using a cell with a path length of 1 mm, bandwidth of 2 nm, response time of 1 s and five accumulations, and corrected for buffer baseline contribution.
Results
Development and analysis of initial structures
Homology modelling
Homology/comparative modelling was attempted, although due to overall very low sequence identity and low homology of the GPA-ECD to known 3D structures of proteins (both solved by crystallography as well as NMR as reported in the Protein Data Bank (PDB) [Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB), RRID:SCR_012820)] this method was not suitable. Partial matches obtained by performing BLAST (29) (NCBI BLAST, RRID:SCR_004870) analysis against the PDB database revealed fragments of 20-30 residues in length from GPYA-EC exhibiting a sequence identity of 38–50% with known structures. However, homology modelling techniques require a minimum of approx. 150 residues with homology of at least 30–50% (30,31). As all the returned sequences were so short, it is unlikely that they reflect the appropriate secondary/tertiary structure as they are missing numerous interactions with other surrounding structures. Most of these segments were helical in nature (Table S2), but were too small to build a reliable homology model.
Threading—iTasser and FALCON
As homology modelling was an inappropriate method for determining a suitable starting structure, the threading method (iTasser) was implemented for the structure prediction for GPA-ECD. The fold prediction is made by “threading” each amino acid in the target sequence to a position in the top 10 template structures generated from the LOMETS threading programs. For each target, iTasser uses the SPICKER program to cluster large ensembles of structural conformations based on pair-wise structure similarity and selects the top 5 models, where the confidence of each is measured by a C-score (32-40). However, this method relies on the fact that the total number of protein folds in nature is much smaller than the total number of known sequences (41,42).
None of the 5 models of GPA-ECD produced by iTasser had positive or relatively high C-scores, indicating a low level of confidence. Visual analysis using PyMOL and VMD revealed these 5 models mostly exhibited a random-coil structure, in contrast to comparative modelling of short GPA sequence segments (approx. 25 amino acids long), where all results returned as α-helical structures.
Subsequently, an alternative threading method, FALCON was used. FALCON uses homologous templates to calculate common structural frameworks using the TBM (Template Based Modelling, including homology and threading) module. If the TBM module fails to identify protein homologues, the ab initio module is activated. Initial attempts for GPA-ECD led to generation of 10 structural models, but none had confidence levels that justified continuation of study.
A previous model had been created using the FALCON program (15), so that program was also adopted for the current work. The previous model was created using residues 21–91 (71 residues), as compared to 19–89 (71 residues) used in the current study. The previously created structure consisted of 5 highly coiled, antiparallel β sheets in 1-2-3-5-4-1 topology, referred to as an OB-fold (oligonucleotide/oligosaccharide-binding fold). In our attempts to replicate the previous model using FALCON, the β-barrel structure was obtained only as 10th best-ranked model. Additionally, this model was only possible to obtain using the amino acid residues 21–91, where no β-barrel structures appeared using residues 19–89. Notwithstanding this, our produced replicate model was renamed ‘Structure 6’ and included as one of the selected models for further analysis. However, the sensitivity of the threading method towards a small addition/deletion of residues on the end-regions was surprising, and more investigations may be needed (beyond the scope of the current work) to support this observation.
Ab Initio modelling—Robetta
Due to the divergent structural observations obtained from the homology modelling and threading methods, prediction of the 3D structure of GPA-ECD was attempted using the program Robetta that implements ab initio methods. The ab initio Relax application consists of two main steps. The first step is a coarse-grained fragment-based search through conformational space using a knowledge-based “centroid” score function that favours protein-like features. The second optional step is all-atom refinement using the Robetta full-atom force field (43). Robetta was selected due to its success in the bi-annual “Critical Assessment of Protein Structure Prediction” (CASP) (44) experiments, which test the latest methods for protein structure prediction. The top 5 models obtained from Robetta were renamed as Structures 1–5 respectively and chosen for further investigation. When selecting models, weight was given not only to the ranking of the model by the Robetta program, but also to the expectation that residues involved in glycosylation were exposed to the surface.
Analysis of GPA-ECD structures
All six initial predicted structures (5 from Robetta and 1 from FALCON) exhibited distinct secondary structural features in the form of 3 to 6 β-sheets (listed in Table S3). There was a consistent feature of two β-sheets across all models, where the first β-sheet was located within the region 64–71 [EISVRTVY] and the second was located within the region 80–87 [RVQLAHHF] (Table S3). Although each β-sheet differed slightly in length and location, these β-sheets were also retained across all models throughout the simulation using the GROMOS force field. The combined region 64–85 is highlighted in red in Figure 1 as a comparison between the initial structure predictions. This β-hairpin is also the site of a number of GPA antigens (Table S1). Another region of consensus was a β-sheet found to span the region 25–30 [VAMHTS] (Table S3). However, the β-sheet identified at 25–30 disappeared within 20 ns of MD simulation and was thus not considered a stable β-sheet region. Using secondary structure prediction program PsiPred (45) short β-sheets were predicted to be located in corresponding location 64-71 [EISVRTVY] (Figure S1). Despite the consensus of the three aforementioned β-sheets, the structure for the entire GPA-ECD of the initial predictions varies greatly, and is likely to be associated with the inherent uncertainties in predicting secondary structure from primary sequence alone (46,47).
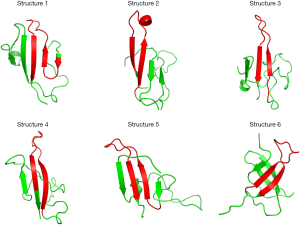
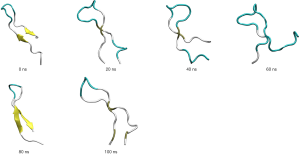
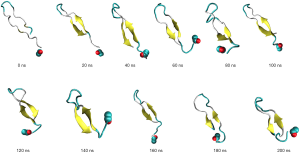
MD simulations were performed in explicit water on the six initial structures to validate the stability of these secondary structures. Furthermore, as the initial structures differed from one another, MD was used to determine if two or more would converge into a single stable representative form. After 200 ns of MD in explicit solvent, visual analysis indicated that all models were unable to retain their secondary structure, failed to conform into a single common structure, and all tended towards complete unfolding of the proteins captured at 0, 100 and 200 ns (Figure 2).
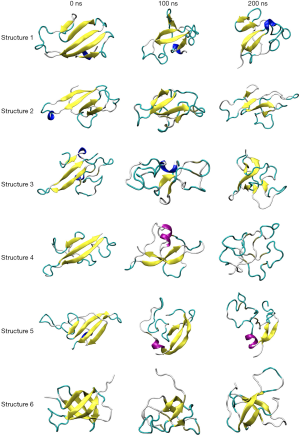
For Structures 1, 2 and 3, a notable feature was the lengthening or shortening of β-sheets as well as significant twisting of the β-sheets seen by visual analysis (Figure 2). This distortion of shape correlates with the fluctuating RMSD values shown in the Y-axis of Figure S2 for Structures 1, 2 and 3. A stable conformation was not found for Structure 4 as it completely unfolds to form a random-coil structure after 200 ns of MD simulation (Figure 2), also illustrated in Figure S2. The RMSD value of Structure 4 continuously increases from 0.3 to 0.6 nm over the entire 200 ns simulation, indicating continuous protein backbone alteration. Structure 5 had more widely fluctuating RMSD values (Figure S2, where the changes in secondary structure are visible across Figure 2). Structure 6 presented with the highest level of secondary structure stability. Although the RMSD values varied within the first 50 ns between 0.3 and 0.6 nm (Figure S2), indicating an unstable starting structure [most likely the alteration of the β-sheet at 27–30 (MHTS)], then at 50 ns there was a sharp spike in RMSD to 0.65 nm indicating a transition in the secondary structure, which remains stable at 0.6–0.65 nm for the rest of the 200 ns simulation. This revealed that a reasonably stable structure has been obtained, which is corroborated by visual inspection of the structure at each time point shown in Figure 2. The RMSF was also calculated for each of the protein structures (Figure S3) where Structures 1, 2, 4 and 5 show high levels of fluctuations throughout the protein. Structures 3 and 5 appear to have lower levels of fluctuation, with the exception of two peaks both between residues 40 to 50, and 70 to 80, as well as high fluctuation on the termini of the protein. However, under visual analysis (Figure 2) it can be seen that the secondary structural features shift from β-sheets to random coils, as well as changes in the tertiary structure.
Across all six structures, the twisting of β-sheets and presence of transient α-helical segments (Figure 2) suggests a propensity to shift into an α-helical structure. However, the majority of observed α-helical segments appeared intermittently at different locations across the six structures as well as at different time-points during the simulation (Table S4). The majority of the helices produced were also 3–10 helices which have been proposed to be an intermediate conformation, although four (of twelve) observed helices were not 3–10 helices. Lastly, the α-helical segments were on average 3 amino acids long and thus were not considered to be a significant structural feature even if they were retained for the majority of the 200 ns simulation.
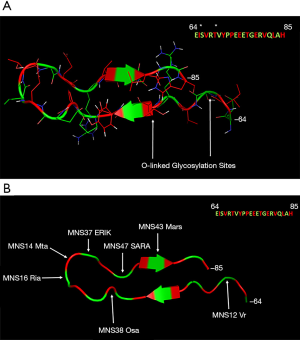
Although the overall secondary structure failed to find a stable form, there were two stable β-sheets. The β-sheet at 81–87 [VQLAHHF] was retained throughout the 200 ns MD simulation for Structures 1,2,3,5 and 6, as well as for more than 100 ns in Structure 4. This was also observed for the β-sheet at 64–67 [EISV] (with the exception of Structure 3). These two β-sheets are located in the exon 3–4 junction region. These sheets are present in the same location as in the initial Robetta and FALCON predicted structures (Figure 1 in red), indicating that these β-sheets may be a stable component of the GPA-ECD. A representative structure of the exon 3–4 junction region was obtained from the combined trajectories of 1 µs of MD simulation, this structure exhibited a β-hairpin-like structure with an internal protein backbone, with side chains and glycosylation sites exposed externally (Figure 3A). This structure is also the location for seven GPA antigens which have been identified on Figure 3B and Table S1.
In relation to the addition of counter-ions, there is currently no strong evidence that neutralisation of the protein enhances stabilisation, and is highly dependent on the model variant and procedure used (48). However, a test for MD simulation of Structure 1 was performed in a 100 mM NaCl environment and run for 100 ns in explicit water, where no additional stability was seen.
GPA-ECD as an intrinsically disordered-like protein
Considering the size of the GPA protein, four independent approaches were used (ab initio, threading, homology modelling and MD simulations) to derive a consensus about the secondary structure of its extracellular domain. The results of the MD simulations across the 6 structures lead to the observation that monomeric form of GPA-ECD most likely exists in an intrinsically disordered form with a small predicted region of local stability of the β-hairpin-like structure across the exon 3–4 junction.
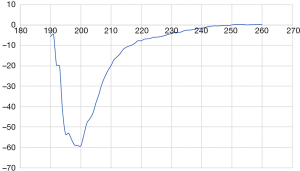
In addition, experiments were performed to test if GPA-ECD is disordered, using circular dichroism (CD) spectropolarimetry. A non-glycosylated peptide of the GPA-ECD was analysed and the CD spectrum indicated that the peptide does not have significant α-helical or β-sheet secondary structure elements and it is most likely in a random coil conformation in solution (Figure 4).
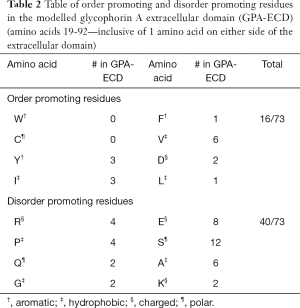
Another method of predicting IDPs is through sequence analysis (49). This follows the principle that IDPs have lower frequencies of order-promoting residues (W, C, I, F, D and L) and hydrophobic/aromatic residues, as well as higher frequencies of disorder-promoting residues (R, P, Q, G, E, S, A and K) and charged/polar residues (50,51). This pattern can be seen in the GPA-ECD (as shown in Table 2), where over half of the residues within the extracellular domain are disorder promoting, and only approximately one fifth are order promoting. Lastly, secondary structure prediction based on the entire GPA protein sequence using PsiPred and DISOPRED (45,52) predicted disordered regions predominantly in exon 2 and 4 (Figure S1).
Full table
Antigen presentation in GPA-ECD
The M/N (MNS1/2), MNTD (MNS46) and Nya (MNS18) antigens were located in regions on the GPA-ECD that were highly unstable and had no consensus for secondary structure across the six models after MD simulation. As a result, no conclusive data could be determined for secondary structure prediction of these antigenic sites.
The exon 3–4 junction was the only region of stability across the six models, and is the location for seven MNS antigens encoded by single amino acid changes (Table S1 and Figure 3B). To determine whether these alterations would reduce the stability of the β-hairpin-like structure, or allow the formation of new secondary structures, seven new structures were produced. Each new β-hairpin-like structure contained a single amino acid change corresponding to one of the seven MNS antigens (Table S1). It was found that after 100 ns of MD simulation, only one single amino acid variation caused disruption to the β-hairpin structure; p.Gly78Arg (MNS:37 ERIK+) (Figure 5). Although slight alterations were noted in p.Ser66Tyr (MNS:12 Vr+), p.Pro73Ser [MNS:38 Os(a+)] and p.Gly82Lys (MNS:−42,43 MARS+), by the end of the 100 ns simulation the β-hairpin-like structure had re-formed. For p.Glu76Lys [MNS:16 Ri(a+)], p.Thr77Ile [MNS:14 Mt(a+)] and p.Arg80Ser (MNS:47 SARA+), the core β-hairpin-like structure was maintained. Additionally, no single amino acid changes were able to produce a stable alternative structure to the β-hairpin. This indicates that only the amino acid change associated with the ERIK antigen causes a structural change in GPA-ECD, and this would be expected to have an impact on the presentation of the antigen for antibody production. It should also be noted that p.Ser66Tyr (MNS:12 Vr+) mutation is also an O-linked glycosylation site, and the alteration of the Serine to a Tyrosine would result in the elimination of this glycosylation site, which would be expected to change the surface presentation of GPA-ECD.
Development of suitable peptides mimicking the β-hairpin-like structure of the exon 3–4 junction region
The coordinates of the representative hairpin structure obtained from the 1 µs MD simulation of GPA-ECD were used to extract a shorter (22 amino acid) peptide sequence that was used for further computational analysis. The hairpin peptide ran for 200 ns MD simulation and was able to retain its secondary structure despite slight extension and shortening of the β-sheet (shown in Figure 6). As peptide synthesis is most commonly carried out linearly, the extended peptide structure (with no pre-determined structure) was also tested to determine if a linear peptide would be able to fold into the β-hairpin-like structure.
MD simulations starting with the linear peptide, in contrast to the simulations carried out from the hairpin structure, were unable to fold into the β-hairpin-like structure or conform into a single stable structure after 200 ns of MD simulations (shown in Figure 7).
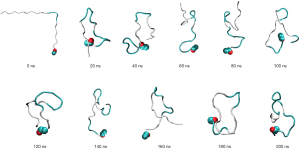
As the linear peptide was deemed to be an unsuitable candidate for laboratory experimental purposes, it was hypothesised that a circular peptide might replicate the modelled β-hairpin-like structure without the need for a protein scaffold. The modelled circular peptide had limited ability to retain the β-hairpin-like structure, as can be seen in Figure 8. Although the circular peptide was able to retain a hairpin-like structure for the first 80 ns, initial twisting can be seen at 120 ns, as well as further folding and collapsing of the circular peptide from 160 ns onward. With a shortened circular peptide length this twisting and folding may reduce.
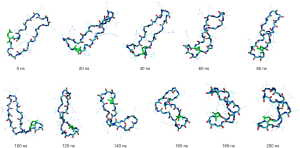
When starting from a β-hairpin structure, or within the confines of the entire GPA-ECD, the β-hairpin peptide was stable. However, peptides that did not start as a β-hairpin (starting from a circular or linear peptide) were unable to re-form the β-hairpin-like structure after 100 ns MD.
Discussion
From the initial searches for homologous protein structures, it was assumed that our predicted structure would similarly contain α-helices. However this was not the case across any of our models. This is likely due to the predicted behaviour of short peptide fragments (20–30 amino acids) not necessarily being replicated in a large molecular structure. In particular, that the formation of β-sheets relies heavily upon tertiary structure and cannot be replicated in smaller fragments (53). As secondary structure is determined by a multiplicity of molecular interactions outside the short domain subjected to sequence comparison, it is not surprising that the short sequences of homology did not align with the produced models.
A key setback in our initial models is the lack of glycosylation. There is evidence that glycosylation might not alter the structure and folding of proteins, but rather acts to enhance protein stability via destabilizing the unfolded protein state (54,55). Alternatively, glycosylation removal allows for recognition of misfolded glycoproteins for proteasome degradation (56). Additionally, a previous study by Ekman et al. [2019] using a similar structure to Structure 6 indicated that the addition of glycosylation (substituting full glycosylation for α-N-Acetyl-D-galactosamine molecules) had a slight stabilization effect but did not play a major role in the determination of GPA-ECD structure (15). On this basis, it is unlikely that the lack of glycosylation is the cause of the disordered nature of the simulated structures. As a result, there was sufficient confidence in the ability to produce accurate secondary structures using the protein backbone and in silico methods without glycosylation, with the intention to add glycosylation once the base structure had been determined.
Although the programs used in this study were deemed suitable in the production of a protein backbone, both Robetta and FALCON have been developed and tested against ordered structures (12,14,57). Thus, challenges arise when attempting to model structures with higher levels of disorder. To overcome these challenges, MD simulations are used in adjunct to such predictive programs and allow for the “settling” of the predicted structures into energetically favourable and hence more probable states. The necessity of the use of MD can be seen when comparing the extracted Exon 3–4 region to its counterpart from the initial protein predictions (highlighted in red in Figure 1), as the β-strands are slightly altered. Furthermore, investigation needs to be undertaken to understand why the Exon 3–4 β-strands were retained whereas the other predicted β-strands disappeared over the 200 ns of MD simulation.
The CD spectropolarimetry results support the hypothesis that the monomeric non-glycosylated GPA-ECD has a high level of disorder, yet seem to contradict the presence of a stable β-hairpin within the GPA monomer. However, it should be noted that although CD spectropolarimetry, works well for proteins that are completely composed of α-helices and to a lesser extent completely composed of β-strands, it has been suggested that CD spectra is not effective at distinguishing mixed α-helical or β-sheet elements with particular difficulty identifying non-canonical β-strands (58). It is highly likely that the stable β-hairpin observed in our computer models was obscured in the CD experiments.
The combination of CD spectropolarimetry results, sequence based prediction, and the visual results of the MD simulations supports the theory that the monomeric extracellular region of GPA has a high level of disorder. As IDPs are able to conform into different 3D-structures, allowing them to interact with a variety of different proteins (51), the intrinsically disordered-like character of monomeric GPA-ECD could explain GPA’s ability to interact with numerous RBC surface proteins. An important feature of GPA is its ability to form stable homodimers (59), but this is not inconsistent with the possibility that the monomeric GPA-ECD is IDP-like. IDP monomers are known to form stable homodimers through specific interactions (60,61). As GPA dimerisation is facilitated via transmembrane interactions (8), it is possible that membrane driven dimerisation stabilises the extracellular domain of the monomer.
When attempting to identify antigenic sites, or develop antibodies against the GPA-ECD, the monomeric form of this molecule may not be a suitable representation of antigens in the context of an RBC. It is likely that antigen presentation in GPA-ECD is comprised of a combination of features including specific amino acid sequences, orientation, and presence of glycosylation. Consideration should also be made to the fact that the process of dimerisation may affect ECD antigen presentation. Additionally, GPA has direct interaction with, and exists in a complex alongside other RBC membrane proteins (62,63). These interactions may also play a role in the tertiary structural presentation of the ECD, as well as antigen presentation such as the case of the Wrb antigen (64). Nonetheless, the stable β-hairpin-like structure that was evident in the MD simulations may be used to improve antigen presentation of the exon 3–4 junction region for future laboratory-based experiments.
The models of a peptide structure of the β-hairpin indicates that the hairpin structure is stable even when removed from the influence of the neighbouring amino acids. This further implies that if the hairpin structure is synthetically produced as a peptide in the β-hairpin-like form it should retain its structure and thus be a useful embodiment of the native structure.
The inability of the circular or extended peptide to fold into a β-hair pin might have occurred due to an activation energy barrier that restricts the “foldability” of this structure. This energy barrier may not be overcome within the time-scale of MD simulations attempted in the present study. Running the peptide simulation for a longer time-scale or altering the parameters of the simulation to overcome the activation energy barrier may allow for the folding of the peptide.
Another possibility is that the formation of a hairpin structure requires the stabilisation energy of other interactions within the extracellular domain, and hence the isolated linear and circular peptides were unable to fold into the β-hairpin-like state. This inability to form the correct secondary structure from a linear or circular peptide has implications in experiments using synthetic peptides as replicas for antigens such as in epitope mapping and biopanning.
Despite the inability of the modelled circular and linear peptides to form the β-hairpin structure, circular or linear peptides are deemed to be preferred starting structures. This is as neither requires a protein scaffold which would increase the ease of laboratory-based experiments for this antigenic region. Further testing may be performed utilising differing sequence lengths (both lengthening and shortening), to determine an optimum sequence for peptides that can better replicate the β-hairpin-like structure of the exon 3–4 junction region. In addition, the use of other peptide presentation systems needs to be investigated to re-create the putative native secondary structure.
The ultimate aim of this work is to understand the antigenicity associated with structural elements of GPA and in the future glycophorin B (GPB) and hybrid glycophorins. Although glycosylation might not play a significant role in protein structure, its presence is crucial in certain antigen presentation. Thus although some information can be gleaned regarding antigenicity from structural models alone, additional detail still needs to be added to these models to fully understand antigen presentation and antibody recognition. As GPA has such high levels of glycosylation the addition of glycosylation requires computing capabilities beyond what was available for this project. The specific effects of glycosylation on antibody recognition and antigen presentation, will only be determinable once an appropriate protein structure for GPA-ECD has been determined and specific glycosylation has been added to the proposed structure based on experimental findings. Future studies adding glycosylation to GPA would also need to account for the complexity associated with differing styles of glycosylation across different cell lines (65), particularly if GPA is expressed on alternative (non-erythrocyte) cell lines for experimental purposes.
Additional studies, both computation and experimental will be required to validate the predicted extracellular structure for GPA, as well as the production of GPA homodimers. Once it can be determined that this modelling process is able to produce suitably accurate extracellular representations in silico, further models can be produced. GPA could be used as a template for homology modelling of GPB and other hybrid glycophorins due to their high level of homology (4). The model produced could also act as a template for the development of a GPA-ECD homodimer model.
Conclusions
In conclusion, this study predicts that the monomeric extracellular domain of non-glycosylated GPA has a high level of disorder; with the exception of the antigenic region that spans the exon 3–4 junction (residues 64–85), and presents as two β-sheets flanking a loop in a hairpin-like structure. Although this structure is locally stable within a simulation of the entire GPA extracellular domain, when starting from a linear or circular peptide, it failed to fold into the β-hairpin structure during the time-scale used in the current MD simulation study. This suggests that stabilisation of the β-sheet-hairpin secondary structure depends upon interactions with other regions of the extracellular domain that are brought into proximity during folding of the tertiary structure. As such, laboratory based experiments to replicate this region will require careful consideration of both secondary and tertiary structure. Lastly, it was shown that after 100 ns MD simulation on the mutated hairpin peptide, the only single amino acid variation, p.Gly78Arg (the ERIK antigen of the MNS system) led to the loss of the β-hairpin secondary structure.
Acknowledgments
Computational resources were provided by the Research Computing Centre (RCC) at the University of Queensland (UQ). All simulations were performed on the Wiener (GPU) cluster. The authors would like to thank Assoc. Prof. Mehdi Mobli and Centre for Advanced Imaging (CAI) at UQ for their assistance with performing the Circular Dichroism experiment.
Funding: Funding for the project was provided from the ARC Training Centre for Biopharmaceutical Innovation (CBI) from the Australian Research Council (ARC), and the Australian Red Cross Lifeblood. Australian Governments fund Australian Red Cross Lifeblood for the provision of blood, blood products and services to the Australian community.
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/aob-20-51
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aob-20-51). Prof. RF serves as an unpaid editorial board member of Annals of Blood. The other authors have no conflicts of interest to declare.
Ethical Statement: the authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Poole J, Daniels G. Blood Group Antibodies and Their Significance in Transfusion Medicine. Transfus Med Rev 2007;21:58-71. [Crossref] [PubMed]
- Heathcote DJ, Carroll TE, Flower RL. Sixty Years of Antibodies to MNS System Hybrid Glycophorins: What Have We Learned? Transfus Med Rev 2011;25:111-24. [Crossref] [PubMed]
- Pisano A, Redmond JW, Williams KL, et al. Glycosylation sites identified by solid-phase Edman degradation: O-linked glycosylation motifs on human glycophorin A. Glycobiology 1993;3:429-35. [Crossref] [PubMed]
- Daniels G. MNS Blood Group System. In: Daniels G. editor. Human Blood Groups. Victoria: Blackwell Science Asia Pty; 2007.
- Chang VT, Crispin M, Aricescu AR, et al. Glycoprotein structural genomics: solving the glycosylation problem. Structure 2007;15:267-73. [Crossref] [PubMed]
- Lee JE, Fusco ML, Saphire EO. An efficient platform for screening expression and crystallization of glycoproteins produced in human cells. Nat Protoc 2009;4:592. [Crossref] [PubMed]
- Stura EA, Nemerow GR, Wilson IA. Strategies in the crystallization of glycoproteins and protein complexes. J Cryst Growth 1992;122:273-85. [Crossref]
- MacKenzie KR, Prestegard JH, Engelman DM. A Transmembrane Helix Dimer: Structure and Implications. Science 1997;276:131-3. [Crossref] [PubMed]
- Schulte TH, Marchesi VT. Conformation of Human Erythrocyte Glycophorin A and Its Constituent Peptides. Biochemistry 1979;18:275-80. [Crossref] [PubMed]
- Dill K, Hu S, Berman E, et al. One- and two-dimensional NMR studies of the N-terminal portion of glycophorin A at 11.7 Tesla. J Protein Chem 1990;9:129-36. [Crossref] [PubMed]
- Consortium TU. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 2019;47:D506-15. [Crossref] [PubMed]
- Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res 2004;32:W526-31 [Crossref] [PubMed]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 2008;9:40. [Crossref] [PubMed]
- Wang C, Zhang H, Zheng WM, et al. FALCON@home: a high-throughput protein structure prediction server based on remote homologue recognition. Bioinformatics 2016;32:462-4. [Crossref] [PubMed]
- Ekman S, Flower R, Hyland C, et al. In silico model for Glycophorin A (GPA) structure. Pathology 2019;51:S123. [Crossref]
- DeLano WL. Pymol: An open-source moleuclar graphics tool. CCP4 Newsletter On Protein Crystallography 2002;40:82-92.
- Christen M, Hünenberger PH, Bakowies D, et al. The GROMOS software for biomolecular simulation: GROMOS05. J Comput Chem 2005;26:1719-51. [Crossref] [PubMed]
- Schymkowitz J, Borg J, Stricher F, et al. The FoldX web server: an online force field. Nucleic Acids Res 2005;33:W382-8 [Crossref] [PubMed]
- Abraham MJ, Murtola T, Schulz R, et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015;1-2:19-25. [Crossref]
- Oostenbrink C, Villa A, Mark AE, et al. A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6. J Comput Chem 2004;25:1656-76. [Crossref] [PubMed]
- Schmid N, Eichenberger AP, Choutko A, et al. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur Biophys J 2011;40:843. [Crossref] [PubMed]
- Berendsen HJC, Postma JPM, van Gunsteren WF, et al. Interaction Models for Water in Relation to Protein Hydration. In: Pullman B, editor. Intermolecular Forces: Proceedings of the Fourteenth Jerusalem Symposium on Quantum Chemistry and Biochemistry Held in Jerusalem, Israel, April 13–16, 1981. Dordrecht: Springer Netherlands; 1981:331-42.
- Berendsen HJC, Postma JPM. Molecular dynamics with coupling to an external bath. J Chem Phys 1984;81:3684-90. [Crossref]
- van Gunsteren WF, Billeter SR, Eising AA, et al. Biomolecular Simulation: The GROMOS96 Manual and User Guide. Biomos; Zürich; 1996.
- Hess B, Bekker H, Berendsen HJC, et al. LINCS: A linear constraint solver for molecular simulations. J Comput Chem 1997;18:1463-72. [Crossref]
- Miyamoto S, Kollman PA. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem 1992;13:952-62. [Crossref]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph 1996;14:33-8, 27-8.
- Daura X, van Gunsteren WF, Mark AE. Folding-unfolding thermodynamics of a β-heptapeptide from equilibrium simulations. Proteins 1999;34:269-80. [Crossref] [PubMed]
- Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol 1990;215:403-10. [Crossref] [PubMed]
- Haddad Y, Adam V, Heger Z. Ten quick tips for homology modeling of high-resolution protein 3D structures. PLoS Comput Biol 2020;16:e1007449 [Crossref] [PubMed]
- Vyas VK, Ukawala RD, Ghate M, et al. Homology modeling a fast tool for drug discovery: current perspectives. Indian J Pharm Sci 2012;74:1-17. [Crossref] [PubMed]
- Wu S, Zhang Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res 2007;35:3375-82. [Crossref] [PubMed]
- Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on a genomic scale. Proc Natl Acad Sci U S A 2004;101:7594-9. [Crossref] [PubMed]
- Karplus K, Barrett C, Hughey R. Hidden Markov models for detecting remote protein homologies. Bioinformatics 1998;14:846-56. [Crossref] [PubMed]
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 1970;48:443-53. [Crossref] [PubMed]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol 1981;147:195-7. [Crossref] [PubMed]
- Zhang Y, Kihara D, Skolnick J. Local energy landscape flattening: Parallel hyperbolic Monte Carlo sampling of protein folding. Proteins 2002;48:192-201. [Crossref] [PubMed]
- Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res 2005;33:2302-9. [Crossref] [PubMed]
- Feig M, Rotkiewicz P, Kolinski A, et al. Accurate reconstruction of all-atom protein representations from side-chain-based low-resolution models. Proteins 2000;41:86-97. [Crossref] [PubMed]
- Canutescu AA, Shelenkov AA, Dunbrack RL Jr. A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci 2003;12:2001-14. [Crossref] [PubMed]
- Grant A, Lee D, Orengo C. Progress towards mapping the universe of protein folds. Genome Biol 2004;5:107. [Crossref] [PubMed]
- Schaeffer RD, Daggett V. Protein folds and protein folding. Protein Eng Des Sel 2011;24:11-9. [Crossref] [PubMed]
- Bradley P, Misura KMS, Baker D. Toward High-Resolution de Novo Structure Prediction for Small Proteins. Science 2005;309:1868-71. [Crossref] [PubMed]
- Kryshtafovych A, Monastyrskyy B, Fidelis K. CASP11 statistics and the prediction center evaluation system. Proteins 2016;84:15-9. [Crossref] [PubMed]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 1999;292:195-202. [Crossref] [PubMed]
- Bywater RP. Protein folding: a problem with multiple solutions. J Biomol Struct Dyn 2013;31:351-62. [Crossref] [PubMed]
- Bywater RP. Prediction of protein structural features from sequence data based on Shannon entropy and Kolmogorov complexity. PLoS One 2015;10:e0119306 [Crossref] [PubMed]
- Drabik P, Liwo A, Czaplewski C, et al. The investigation of the effects of counterions in protein dynamics simulations. Protein Eng 2001;14:747-52. [Crossref] [PubMed]
- Oldfield CJ, Dunker AK. Intrinsically Disordered Proteins and Intrinsically Disordered Protein Regions. Annu Rev Biochem 2014;83:553-84. [Crossref] [PubMed]
- Uversky VN. Intrinsically Disordered Proteins and Their “Mysterious” (Meta)Physics. Front Phys 2019; [Crossref]
- Dyson HJ. Making Sense of Intrinsically Disordered Proteins. Biophysical journal 2016;110:1013-6. [Crossref] [PubMed]
- Jones DT, Cozzetto D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics 2015;31:857-63. [Crossref] [PubMed]
- Schneider JP, Kelly JW. Templates That Induce. alpha.-Helical, .beta.-Sheet, and Loop Conformations. Chem Rev 1995;95:2169-87. [Crossref]
- Shental-Bechor D, Levy Y. Effect of glycosylation on protein folding: A close look at thermodynamic stabilization. Proc Natl Acad Sci U S A 2008;105:8256-61. [Crossref] [PubMed]
- Mitra N, Sinha S, Ramya TNC, et al. N-linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem Sci 2006;31:156-63. [Crossref] [PubMed]
- Parodi AJ. Role of N-oligosaccharide endoplasmic reticulum processing reactions in glycoprotein folding and degradation. Biochem J 2000;348:1-13. [Crossref] [PubMed]
- Protein Structure Prediction Center. The Critical Assessment of protein Structure Prediction (CASP). University of California. 2020. Available online: https://predictioncenter.org/index.cgi
- Khrapunov S. Circular dichroism spectroscopy has intrinsic limitations for protein secondary structure analysis. Anal Biochem 2009;389:174-6. [Crossref] [PubMed]
- Gerber D, Shai Y. In Vivo Detection of Hetero-association of Glycophorin-A and Its Mutants within the Membrane. J Biol Chem 2001;276:31229-32. [Crossref] [PubMed]
- Sigalov AB. Unusual biophysics of immune signaling-related intrinsically disordered proteins. Self Nonself 2010;1:271-81. [Crossref] [PubMed]
- Danielsson J, Liljedahl L, Bárány-Wallje E, et al. The intrinsically disordered RNR inhibitor Sml1 is a dynamic dimer. Biochemistry 2008;47:13428-37. [Crossref] [PubMed]
- Williamson RC, Toye AM, Glycophorin A. Band 3 aid. Blood Cells Mol Dis 2008;41:35-43. [Crossref] [PubMed]
- Mankelow TJ, Satchwell TJ, Burton NM. Refined views of multi-protein complexes in the erythrocyte membrane. Blood Cells Mol Dis 2012;49:1-10. [Crossref] [PubMed]
- Poole J. Red cell antigens on band 3 and glycophorin A. Blood Rev 2000;14:31-43. [Crossref] [PubMed]
- Croset A, Delafosse L, Gaudry JP, et al. Differences in the glycosylation of recombinant proteins expressed in HEK and CHO cells. J Biotechnol 2012;161:336-48. [Crossref] [PubMed]
Cite this article as: Ekman S, Flower R, Mahler S, Gould A, Barnard RT, Hyland C, Jones M, Malde AK, Bui XT. In silico molecular dynamics of human glycophorin A (GPA) extracellular structure. Ann Blood 2021;6:11.


