
Single-cell RNA sequencing of peripheral blood mononuclear cells from the patient with acute promyelocytic leukemia: a case study
Acute promyelocytic leukemia (APL), a special type of acute myeloid leukemia (AML, M3 subtype), is an aggressive hematological malignancy (1). APL occurs mostly in the young and middle-aged people, accounting for approximately 10% of the AML cases. It is characterized by the unrestricted proliferation of a large number of leukemia cells in bone marrow and other hematopoietic tissues, with the significantly inhibited hematopoietic function, causing the disease develops rapidly with the dangerous clinical manifestations (2). Bleeding and embolism are prone to occur during the progression of APL, which is the leading cause to the death of APL patients. APL used to be the myeloid leukemia with the highest fatality rate. With the in-depth research on the pathogenesis and therapy of APL, the survival rate of APL patients has been greatly improved under the treatment of retinoic acid (ATRA) and arsenic trioxide (As2O3) (3). However, the early mortality rate is still high since the bleeding and disseminated intravascular coagulation (DIC), the most prominent features of APL, causing the death in the early stage (4). Therefore, to control blood coagulation timely is the key to the treatment in the early stage of APL patients. The early effective intervention is bound to require the rapid and precise diagnosis of APL patients. However, the conventional diagnostic techniques of APL based on the genetics and morphology are still defective and unstable, such as occasional misdiagnosis and time-consuming.
Single-cell RNA sequencing (scRNA-seq) is currently the most advanced technology in the field of precision medicine, with a very broad application prospect in the disease diagnosis (5). This method allows us to quickly and systematically assess the heterogeneity of various types of cells from the patient, identifying the disease-related cells and their genetic and phenotypic characteristics, especially for those few abnormal cells that occur in the early stage of the disease (6). Therefore, this method is particularly suitable for the early detection and companion diagnosis of diseases, such as APL. There have been some studies on the single-cell transcriptomics analysis in AML, but not in APL (7). In this study, we conducted the single-cell transcriptomics detection method based on the 10× Genomics Chromium droplet-based platform to evaluate the single-cell heterogeneity of peripheral blood mononuclear cells (PBMCs) from the APL patient, exploring the feasibility of scRNA-seq for the rapid and precise diagnosis of APL.
The PBMCs was separated from the blood of one APL patient (female, 27 years old) after the patient’s consent and the approval of the ethics committee. This patient presented with skin ecchymosis for 1 month. The complete blood cell count (CBC) revealed was normal white blood cell count (WBC of 9.05×109/L), anemia (hemoglobin of 56 g/L) and thrombocytopenia (platelets 32×109/L). The bone marrow was hyperproliferative (mainly primitive granulocytes and neutrophils), which was consistent with the marrow image of AML. The single-cell transcriptomics research method was carried out as previously described (8). The cells that have unique feature counts over 2,500 or less than 200, and the cells that have >5% mitochondrial unique molecular identifier (UMI) counts were filtered out in the process of quality control by using Seurat. The doublets identified by using DoubletFinder were filtered before analysis. The gene markers used in this study by referring to the previous report and the database CellMarker. We also used the SingleR and Seurat to define clusters via differential expressed markers. Dimensionality reduction for visualizing single-cell data was performed based on the uniform manifold approximation and projection (UMAP) by using Seurat (9).
In this study, we performed systematic screening analysis on the single-cell transcriptomes of the PBMCs sampled from the APL patient (5,888 cells) and healthy people (4,000 cells) (Figure 1). A large difference was observed in the single-cell transcriptomic clustering between the PBMCs of APL patient and healthy people (Figure 1A). There were only 8% of the cells from APL patient clustered with the normal lymphocyte features. Other 92% of the cells clustered closely forming a large abnormal cluster, with the increased promyelocytic cells in peripheral blood uniquely found in the APL patient compared with the AML patients and healthy donors in other researches (Figure 1B). The APL related cells were further clustered into 7 main groups (0, 1, 3, 5, 6, 13, 16, Figure 1C). Furthermore, the expression level of the genes related to APL-derived promyelocytes and DIC were highly expressed in these abnormal cell clusters (Figure 1D), suggesting the promyelocytic cells proliferation and DIC occurred (Figure 1E,F). The representative feature genes MPO and ELANE were highly expressed in the cell clusters of APL PBMCs (Figure 1G). These phenomena indicated this APL patient was under a dangerous condition and the effective treatment was urgently needed. In addition, the presence of leukemia stem cells (LSCs) indicated the prognosis evaluation and efficacy monitoring on LSCs is essential (Figure 1D). Moreover, the representative feature gene CD34 was also highly expressed in the cell clusters of APL PBMCs.
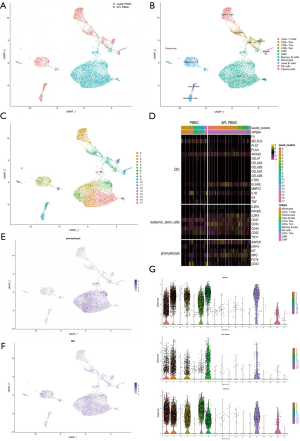
APL is a myeloid proliferative disease characterized by abnormally increased promyelocytic cells in bone marrow and peripheral blood. Conventional APL diagnostic methods include the cell morphology screening for leukemia cells, fluorescence in situ hybridization (FISH) and genetic testing for specific chromosomal translocations t(15;17)(q22;q21), and RT-PCR testing for promyelocytic leukemia-retinoic acid receptor alpha (PML-RARA) fusion genes (10). Cell morphology screening methods require staining and other artificial processing, leading to the morphological changes of cells and further affect the detection results. The accuracy of the results largely depends on the subjective factors and technical level of inspectors, which could cause the occasional misdiagnosis of some atypical patients. The methods of genetic testing on the translocation and fusion gene, usually limited by the number of sample cells with a time lag, only works for a subset of APL patients. Accurate diagnostic typing is an important basis for prompt clinical treatment and prognosis evaluation of APL. Molecular typing system based on scRNA-seq detecting the APL-gene set expression at single-cell resolution is more accurate and comprehensive, providing essential reference for accurate typing, individualized treatment and prognosis evaluation of patients.
By using the single-cell sequencing technology, the features of APL can be easily detected in the early stage. The DIC-related abnormal cells and LSCs could be quickly identified for the prognosis evaluation and efficacy monitoring, which need more research for application. Systematically analyzing the expression of APL related markers, and monitoring tens of thousands of cells at the same time can be effectively implemented with fast speed and high precision. Even a small number of abnormal cells can be also detected effectively in the early stage by using single-cell sequencing to quickly diagnose APL patients can make early detection and prompt interventions for APL patients a reality. It can also be used as a companion diagnostic tool during treatment to accurately assess various risk factors for APL, such as DIC. Nevertheless, the application of this advanced method for APL detection needs to be further improved and the high cost of single-cell sequencing is a challenge remain to be solved.
Acknowledgments
We thank Qianyang Biomedical Research Institute for assistance in sequencing and bioinformatics analysis.
Funding: This study was supported by the National Special Research Program of China for Important Infectious Diseases (2018ZX10302103, 2017ZX10202102-003), the Important Key Program of Natural Science Foundation of China (81730060), National Key Research and Development Program of China (2020YFC0841400), and the Joint-Innovation Program in Healthcare for Special Scientific Research Projects of Guangzhou (201803040002), the China Postdoctoral Science Foundation (2020M683032).
Footnote
Provenance and Peer Review: This article was an unsolicited submission to the journal. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aob-20-65). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang ZY, Chen Z. Acute promyelocytic leukemia: from highly fatal to highly curable. Blood 2008;111:2505-15. [Crossref] [PubMed]
- Hassan IB, Zaabi MRA, Alam A, et al. Characteristics features and factors influencing early death in Acute promyelocytic leukemia; Experience from United Arab Emirates (UAE). Int J Hematol 2017;106:90-8. [Crossref] [PubMed]
- Zhang X, Zhang H, Chen L, et al. Arsenic trioxide and all-trans retinoic acid (ATRA) treatment for acute promyelocytic leukemia in all risk groups: study protocol for a randomized controlled trial. Trials 2018;19:476. [Crossref] [PubMed]
- Chen Z, Wang ZY, Chen SJ. Acute promyelocytic leukemia: cellular and molecular basis of differentiation and apoptosis. Pharmacol Ther. 1997;76:141-9. [Crossref] [PubMed]
- Potter SS. Single-cell RNA sequencing for the study of development, physiology and disease. Nat Rev Nephrol 2018;14:479-92. [Crossref] [PubMed]
- Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol 2018;18:35-45. [Crossref] [PubMed]
- van Galen P, Hovestadt V, Wadsworth Ii MH, et al. Single-Cell RNA-Seq Reveals AML Hierarchies Relevant to Disease Progression and Immunity. Cell 2019;176:1265-1281.e24. [Crossref] [PubMed]
- Zhang J, Liu J, Yuan Y, et al. Two waves of pro-inflammatory factors are released during the influenza A virus (IAV)-driven pulmonary immunopathogenesis. PLoS Pathog 2020;16:e1008334 [Crossref] [PubMed]
- Becht E, McInnes L, Healy J, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 2018;37:38-44. [Crossref] [PubMed]
- Akagi T, Shih LY, Kato M, et al. Hidden abnormalities and novel classification of t(15;17) acute promyelocytic leukemia (APL) based on genomic alterations. Blood 2009;113:1741-8. [Crossref] [PubMed]
Cite this article as: Liu J, Lu L, Liang L, Zhang H, Zhang X. Single-cell RNA sequencing of peripheral blood mononuclear cells from the patient with acute promyelocytic leukemia: a case study. Ann Blood 2021;6:21.


