
Performance evaluation of two supplemental confirmatory assays of hepatitis C virus antibody
Introduction
Hepatitis C virus (HCV) had been found for the first time in 1989 (1). Blood transfusion is still one of the most important transmission route of HCV infection in developing countries up to the present. The epidemic trend of HCV is still not optimistic in China (2). The common HCV genotypes in China are type 1b and 2a, and type 1b accounts for 60% to 70% of those infected with HCV (3,4). In the past 30 years, the detection strategy of HCV infection has evolved from detection of HCV antibody alone to antigens-antibody combined detection with nucleic acid detection, which is widely used currently In China. Enzyme-linked immunosorbent assays (EIA) assay mainly depends on two kinds of domestic anti-HCV EIA reagents or one imported /one domestic reagent according to the “Blood Donation Law”. Beijing Red Cross Blood Center has adopted the latter pattern which has a relatively higher false positive rate (S/CO between 0.7 and 3.8) even though there is a well-known imported reagent. As reported by Warkad et al. that anti-HCV antibodies show high false-positive rates among populations with low (<10%) prevalence of HCV infection (5). Since the EIA reagent cannot distinguish whether the reaction site is directed against the viral protein region or against the non-structural regions (including NS3, NS4, NS5) (6), Chiron developed RIBA 3.0 which is a qualitative immunoblotting strip reagent and had been used as the preferred supplementary serological testing method and gold standard for the diagnosis of HCV infection due to its robust specificity (7).
However, Chiron RIBA 3.0, as the only supplemental anti-HCV test licensed by Food and Drug Administration (FDA), has been permanently discontinued since 2011.A domestic supplemental confirmatory reagent, CWT, has acquired the license from China Food and Drug Administration (CFDA) recently. The manufacturer recommends CWT as supplemental confirmatory regents for blood donors. The epidemic strains of HCV in Mainland China are different from those in Western countries. Therefore, this study intends to preliminarily explore the applicability of CWT in mainland China by comparing the confirmation performance of RIBA 3.0 and CWT on the same batch of specimens.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/aob-20-67).
Methods
Clinical specimens
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Ethical approval for this study was considered unnecessary by the Ethics Committee in our institution (Beijing Red Cross Blood Center) since the “Development of HCV antibody RIBA diagnostic reagent” research project (No. 2008ZX10002-012) has been ethically demonstrated at the time of approval and no patient identifiable data was shared. Informed consent was taken from all the blood donors.
Five hundred and thirty specimens collected from unremunerated blood donors of Beijing Red Cross Blood Center (BRCBC) which were tested anti-HCV reactive by two rounds of EIA according to the screening regulation of National Transfusion Transmitted Infectious Diseases, then the supernatant was taken and stored in a refrigerator at –80 °C separately.
All samples were reactive at least with one kind of EIA kit.
Screening and confirmatory testing
EIA assay
Reagents: The two kinds of anti-HCV EIA assay for these 530 serum specimens were Ortho HCV 3.0, (Ortho Diagnostic Systems, Raritan, NJ, USA) and JWK anti-HCV reagent (Beijing Jin Wei Kai Medical Biotechnology Co., Ltd. Beijing, China) respectively.
Instruments: Specimen processing by Tecan enzyme immunosorbent apparatus (TECAN, Switzerland) and FAME24/30 immunoassay apparatus (HAMILTON, Switzerland).
Supplemental tests of anti-HCV
For RIBA 3.0, the recombinant HCV-encoded antigen and the synthetic HCV-encoded polypeptide have been fixed on the test strip respectively. Among them, Core and NS4 are synthetic antigens, derived from the presumed viral nucleocapsid protein and non-structural protein respectively.NS3 and NS5 and hSOD are recombinant expression antigens derive from E. coli and S. serevisiae (the latter two protein).
For CWT supplemental tests, NS3, NS4-1, NS4-2, NS5, Core protein and two control line proteins were coated on the nitrocellulose strip. Among them, NS3 and control line proteins are recombinant antigens expressed in E. coli. NS4-1, NS4-2, NS5 and Core protein are synthetic proteins. The control lines 1 and 2 must appear at the same time in each experiment. If only one or both of two lines are absent, the experiment is invalid.
The above 530 serum specimens were tested with RIBA 3.0 and CWT at the same time.
Result criteria
In EIA assay, the anti-HCV detection COI ratio >0.7 specimens were retested by double-well. If one well is reactivity the sample was judged as positive, and double- well of non-reactivity were negative.
Supplemental tests of RIBA 3.0 and CWT were operated according to the instruction, and the judgment criteria for the result interpretation for the latter are shown in Tables 1 and 2. The intensity of the colored bands is proportional to the amount of bound antibody and is graded as – (none), ± (indetermination) and 1+ to 2+.

Full table

Full table
Statistical analysis
Statistical analysis was carried out with SPSS statistical software version 22.0 (SPSS Inc., Chicago, IL, USA) for Windows. The consistency of the diagnostic results of HCV3.0 and CWT supplementary experiments uses chi-square test and P-value <0.05 was considered as significantly different and P-value <0.01 was considered as highly significant.
Results
The positive detection rate of CWT and HCV RIBA 3.0
The overall positive rate of the two confirmation reagents
The positive rates of 530 specimens confirmed by CWT and RIBA 3.0 were 34.34% (182/530) and 30.19% (160/530) respectively, and the common positive rates of the two methods were 24.15% (128/530).
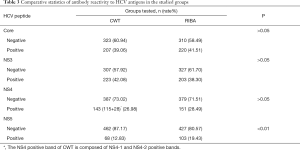
Positive detection results of different HCV peptides
The frequencies of the specific antibody response to the different HCV peptides were studied. The frequency of CWT was NS3, Core, NS4 (NS4-1, NS4-2) and NS5 from strong to weak, while that of RIBA 3.0 was Core, NS3, NS4 and NS5 (Table 3).
Full table
Consistency of CWT and HCV RIBA 3.0 qualitative analysis
Overall consistency of CWT and HCV RIBA 3.0 test results ( Table 4 )

Full table
The diagnostic results of HCV RIBA 3.0 and CWT supplementary experiments are generally consistent (kappa=0.445, P<0.01); And the positive rate of CWT is 34.3%, which is significantly higher than 30.2% of RIBA 3.0, and the difference is highly statistically significant (P<0.01).
Analysis of the number of the positive bands of CWT and RIBA 3.0 reagent
The intensity of the antibody response detected by the number of positive bands observed in RIBA 3.0 and CWT were classified into null band (negative), 1 band (indeterminate), low intensity (2 bands) and high intensity (3–5 bands). The number of low to high intensity antibody bands of CWT and RIBA 3.0 are 182 and 160 respectively. And 64.8% (118/182) specimens were reactive to any three or more of the five antigens of NS3, Core, NS4 (NS4-1, NS4-2) and NS5 in CWT group (high intensity group), while in RIBA 3.0 the reactivity data to three or more of the four antigens (NS3, Core, NS4 and NS5) was 74.4% (119/160) (Table 5).

Full table
The consistency of CWT and RIBA 3.0 different antigen detection
There were 5 specimens that five CWT antigen bands (Core, NS3, NS4-1, NS4-2 and NS5) were all positive and among them 3 specimens were completely corresponding to the specimens of all the four peptides positive in RIBA 3.0 group (No. 488, 1517, 1712). The other 2 specimens for RIBA 3.0 were one positive (two bands positive for Core and NS3, No. 971) and one negative for all the antigen bands (No. 504) (Table 6).

Full table
In 92 specimens with CWT Core, NS3 and NS4-1 antigen positive bands, the corresponding RIBA 3.0 results were 79 cases positive, 8 cases indeterminate (No. 378, 492, 896, 967, 968, 986, 1284 and 1649) and negative in 5 cases (No. 504, 536, 905, 1200 and 1670). In 24 specimens of three antigen bands positive (Core, NS3 and NS4-2) for CWT, that of RIBA 3.0 results were 23 cases positive, and 1 negative (No. 504). In these 23 positive ones, 69.6% specimens (16/23) were all 4 bands of RIBA 3.0 positive. 21.7% (5/23) were 3 bands positive and the other 2 specimens were 2 bands positive.
And among 12 CWT specimens of Core, NS3 and NS5 antigen bands positive, 11 cases were positive for RIBA 3.0 results, and 8 of them (72.7%) were positive for all the four RIBA 3.0 antigen bands, another 2 cases were 3 bands positive and 1 case was 2 bands positive.NO.504 specimen is the only negative one.
Discussion
The residual risk of HCV declined over the last decade due to improved screening reagents, implementation of the nucleic acid amplification test, and tight application of strict donor selection procedures. However, due to China’s unique HCV epidemic strains and certain deficiencies in domestic indirect HCV antibody EIA reagents (the purity of genetically engineered or synthetically prepared fusion protein antigens is insufficient) and the state of blood samples (hemolysis, high IgG concentration, etc.) high false-positive rates among blood donors persists. Supplemental anti-HCV tests are designed to resolve such false-positive testing by EIA. They can help optimize the detection efficiency of anti-HCV reagents and improve the laboratory detection accuracy (conformity rate). On the other hand, they can help to determine whether the donation of blood donors would be delayed indefinitely or recalled again after a certain interval.
We used CWT and RIBA 3.0 supplemental anti-HCV tests to detect 530 EIA positive plasma specimens respectively. The EIA false positive rate of the CWT test group was 65.66% (348/530), and 69.81% (370/530) for RIBA 3.0 group. With the use of supplemental anti-HCV tests, their HCV status is most likely negative with no need for follow up.
Our results showed that 128 specimens were positive for CWT and RIBA 3.0 simultaneously among 530 specimens (24.2%), which is greater than 17.99% (25/139) reported by Chaudhary et al. (8). Except that, 12 specimens were indeterminate for CWT among the remaining 32 RIBA 3.0 positive ones yet 4 of them were positive for all RIBA 3.0 antigen bands (No. 498, 502, 991, 1293). The other 20 cases were CWT negative, yet 5 of them were also positive for all the RIBA 3.0 antigen bands (No. 512, 539, 601, 990, 998). It would be presumed that CWT and RIBA 3.0 may differ in gene sequence selection of HCV antigen bands.
The common indeterminate results were 53 in 530 specimens which accounted for only 32.3% (53/164) in RIBA 3.0 indeterminate specimens and 54.6% (53/97) in those of CWT. Another 36 cases of RIBA 3.0 indeterminate were positive for CWT, among which 20 cases were positive for RIBA 3.0 Core antigen alone, yet among them 18 cases (90%) of the corresponding CWT group were positive for Core/NS3, and the other 2 cases were Core/NS4-1 and NS4-1/NS4-2 positive. Of the 9 individual RIBA 3.0 NS3 antigen positive indeterminate specimens, 5 (55.6%) of the corresponding CWT results were positive for Core/NS3, and 5 were positive for NS3/NS4-1 (No. 986 were positive for four CWT antigen bands except for NS4-2). In the 5 individual RIBA 3.0 NS4 positive indeterminate specimens, 4 cases showed that NS4-1 was co-positive with one or more additional CWT antigen peptides and the last one was CWT Core/NS3 co-positive. The possible causes of this phenomenon should be related to the selection of HCV antigen fragments used by the two supplemental reagents, the gene sequence of coated fragments of the blood testing EIA reagents, the reagents production processes as well as the corresponding detection procedures in addition to HCV geographical and population differences in the distribution of HCV genotypes.
In 44 CWT indeterminate alone specimens, 12 cases (4 specimens were positive for all the RIBA 3.0 antigen bands) were RIBA 3.0 positive and 32 cases (24 cases positive for CWT NS3 and 4 cases positive for Core) were RIBA 3.0 negative. This shows that the detection consistency of the two reagents is not high, each has different focus. The positive detection rate of CWT NS3 antigen peptides in Table 3 was higher than RIBA 3.0 (223:203) which was similar with the report of Yu et al. (9).
One hundred and fifty-two cases of CWT NS3 positive were involved in 203 cases of individual NS3 positive RIBA 3.0 indeterminate samples. Among the remaining 51 specimens, 2 cases (NO. 1076, 1626) were CWT positive for core peptide common with NS4 or NS5 respectively. Five specimens were CWT indeterminate including 4 positive for core peptide and 1 positive for NS5 peptide.NS3 is always used in diagnosis for its highly immunogenic as well as nucleoside triphosphatase and helicase activity. NS3 and NS4-1 form the active, heterodimeric serine protease which is the target of medicinal chemistry efforts (10).
In addition, positive NS3 indicates a greater likelihood of active HCV infection (11). Isolated Core or NS3 reactivity means a higher probability of true presence of anti-HCV antibodies and may be the sign of acute or chronic HCV infection in case of a high S/CO ratio and strong reactivity on the immunoblot assay (12,13). Therefore, high detection ability for NS3 of CWT can better identify active HCV carriers in blood donors.
However, the detection consistency of NS3 between the two reagents is not very high (74.88%, 152/203) which may be related to the difference of target NS3 genetic sequences selection by the two reagents and the different prevalence of HCV genotypes and subtypes in different regions.
The drawback of the third-generation EIA assays was that the low positive predictive values in a low prevalence of HCV infection (<10%) (5) which require confirmation with other more specific supplementary tests. So due to its robust specificity, RIBA had been used as the preferred test for a long period. Even now some researchers reported that RIBA is still necessary for the detection of false positive cases which occur quite frequently in countries of high prevalence (14).
HCV prevalence in the general population was 1.6% in China (15). And Liu et al. reported that the overall change in HCV reporting incidences in China from 2004 to 2014 was 1.16 (95% CI 1.12–1.20, P<0.001), and most provinces exhibited an increasing trend in HCV reporting incidence (2). The prevalence of HCV among first-time donors was 166.56 per 100,000 donors (95% CI, 156.04–177.08 per 100,000 donors) from June 2013 to December 2016 across five China blood centers (16).
Screening tests for antibodies to HCV may generate up to 32% false positivity in low-risk populations (17). Fu et al. advised that HCV donor screening procedures should be improved by incorporating confirmatory testing into routine blood screening in blood center to reduce unnecessary donor loss, which should be a better way to monitor and control the risk of transfusion-transmitted HCV infection at present in China (16).
The low positive rate (24.2%, 128/530) of confirmatory test of EIA screening positive samples in our study suggests that the specificity of anti-HCV EIA reagents for blood screening still needs to be improved. Furthermore, confirmation is valuable for surveillance in the absence of HCV RNA testing (17). The number of positive cases confirmed by CWT is higher than that of RIBA 3.0 (182:160), which also proves the applicability of CWT in China. In fact, for countries with moderately high HCV prevalence, RIBA can still play a “gold standard” in measuring the detection capacity of newly developed HCV EIA reagents and determining inconsistent/ discrepant results (18).
Indeterminate RIBA results could involve instances in which past HCV infections occurred without total elimination of the antibodies (19). Makuria et al. (20) also reported that RIBA indeterminate blood donors were older than spontaneously recovered subjects or chronic HCV carriers. The older age suggests that their HCV exposure might have been in the remote past so that some anti-HCV antibody responses to have waned. The proportion of CWT indeterminate results were a little high (97/530) in our study, but due to consideration of protecting personal privacy, we did not further analyze the age of blood donors.
Except for RIBA, the corroboration of HCV infection can also be confirmed by another confirmatory testing method, HCV-RNA detection. In our experiments, It is regretted that HCV-RNA detection had not been done for the CWT indeterminate specimens due to the volume limitation of part of specimens and deficiency of nucleic acid testing equipment. Pereira et al. (7) reported that 14 blood samples with indeterminate RIBA results had undetectable viral loads (detection limit ≤50 IU/mL). The results are still the same even though 71.4% (10/14) of these samples were reevaluated six months later. So he concluded that individuals with indeterminate RIBA results had no detectable HCV-RNA. However, Pawlotsky et al. reported that 31 (52.5%, 31/59) RIBA 3.0 indeterminate serum samples were HCV RNA positive by PCR (21).
Acknowledgments
The authors thank Beijing Red Cross blood centers for providing the samples.
Funding: This subject was supported by “Development of HCV antibody RIBA diagnostic reagent” research project (No. 2008ZX10002-012).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/aob-20-67
Data Sharing Statement: Available at http://dx.doi.org/10.21037/aob-20-67
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/aob-20-67). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Ethical approval for this study was considered unnecessary by the Ethics Committee in our institution (Beijing Red Cross Blood Center) since the “Development of HCV antibody RIBA diagnostic reagent” research project (No. 2008ZX10002-012) has been ethically demonstrated at the time of approval and no patient identifiable data was shared. Informed consent was taken from all the blood donors.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Allain JP. Hepatitis C virus in blood donation. Lancet 2005;365:276-8. [Crossref] [PubMed]
- Liu Z, Yang Q, Shi O, et al. The epidemiology of hepatitis B and hepatitis C infections in China from 2004 to 2014: an observational population based study. J Viral Hepat 2018;25:1543-54. [Crossref] [PubMed]
- Wu HB, Zhou ZX, Huang YX. Analysis of epidemiological characteristics of viral hepatitis C in China, 1. 2004—2011. Modern Preventive Medicine 2015;7:1173-5.
- Li J, Chen J, Zhuang H. Epidemiology of hepatitis C. Journal of Practical Hepatology 2012;5:379-81.
- Warkad SD, Song KS, Pal D, et al. Developments in the HCV Screening Technologies Based on the Detection of Antigens and Antibodies. Sensors (Basel) 2019;19:4257. [Crossref] [PubMed]
- Van der Poel CL, Cuypers HT, Reesink HW, et al. Confirmation of hepatitis C virus infection by new four-antigen recombinant immunoblot assay. Lancet 1991;337:317-9. [Crossref] [PubMed]
- Pereira FM, Zarife MA, Reis EA, et al. Indeterminate RIBA results were associated with the absence of hepatitis C virus RNA(HCV-RNA) in blood donors. Rev Soc Bras Med Trop 2014;47:12-7. [Crossref] [PubMed]
- Chaudhary RK, MacLean C. Evaluation of first- and second-generation RIBA kits for detection of antibody to hepatitis C virus. J Clin Microbiol 1991;29:2329-30. [Crossref] [PubMed]
- Yu Y, Gu JL, Liang ZHL. Comparison of determination results by three confirmatory kits for hepatitis C virus antibody. Chin J Biologicals 2013;9:1296-9.
- Steinkühler C, Koch U, Narjes F, et al. Hepatitis C virus protease inhibitors: current progress and future challenges. Curr Med Chem 2001;8:919-32. [Crossref] [PubMed]
- Tobler LH, Stramer SL, Chien DY, et al. Antibodies to a novel antigen in acute hepatitis C virus infections. Vox Sang 2007;92:1-7. [Crossref] [PubMed]
- Kiely P, Kay D, Parker S, et al. The significance of third-generation HCV RIBA-indeterminate, RNA-negative results in voluntary blood donors screened with sequential third-generation immunoassays. Transfusion 2004;44:349-58. [Crossref] [PubMed]
- Lemaire JM, Courouce AM, Defer C, et al. HCV RNA in blood donors with isolated reactivities by third-generation RIBA. Transfusion 2000;40:867-70. [Crossref] [PubMed]
- Rafik M, Bakr S, Soliman D, et al. Characterization of differential antibody production against hepatitis C virus in different HCV infection status. Virol J 2016;13:116. [Crossref] [PubMed]
- Bennett H, Waser N, Johnston K, et al. A review of the burden of hepatitis C virus infection in China, Japan, South Korea and Taiwan. Hepatol Int 2015;9:378-90. [Crossref] [PubMed]
- Fu P, Lv Y, Zhang H, et al. Hepatitis C virus prevalence and incidence estimates among Chinese blood donors. Transfusion 2019;59:2913-21. [Crossref] [PubMed]
- Kodani M, Martin M, de Castro VL, et al. An Automated Immunoblot Method for Detection of IgG Antibodies to Hepatitis C Virus: a Potential Supplemental Antibody Confirmatory Assay. J Clin Microbiol 2019;57:e01567-18. [Crossref] [PubMed]
- Hyun J, Ko DH, Kang HJ, et al. Evaluation of the VIDAS Anti-HCV Assay for Detection of Hepatitis C Virus Infection. Ann Lab Med 2016;36:550-4. [Crossref] [PubMed]
- Bes M, Esteban JI, Casamitjana N, et al. Hepatitis C virus (HCV)-specific T-cell responses among recombinant immunoblot assay-3-indeterminate blood donors: a confirmatory evidence of HCV exposure. Transfusion 2009;49:1296-305. [Crossref] [PubMed]
- Makuria AT, Raghuraman S, Burbelo P, et al. The clinical relevance of persistent recombinant immunoblot assay-indeterminate reactions: insights into the natural history of hepatitis C virus infection and implications for donor counseling. Transfusion 2012;52:1940-8. [Crossref] [PubMed]
- Pawlotsky JM, Bastie A, Pellet C, et al. Significance of indeterminate third-generation hepatitis C virus recombinant immunoblot assay. J Clin Microbiol 1996;34:80-3. [Crossref] [PubMed]
Cite this article as: Zhang L, Zha Y, Shi L, Qiu Y. Performance evaluation of two supplemental confirmatory assays of hepatitis C virus antibody. Ann Blood 2021;6:12.


