
Elucidation of the molecular bases of the Rh system and its contribution to transfusion and obstetric medicine—historical and current perspective: a review
Introduction
The Rh system is of major clinical interest in transfusion and obstetric medicine due to the involvement of Rh antibodies in phenomena of immune-mediated erythrocytes destruction. The five principal Rh antigens—D, C, c, E, and e—are highly immunogenic and play a central role in the pathogenesis of the Haemolytic Disease of the Newborn (HDN), haemolytic transfusion reactions and some cases of autoimmune haemolytic anaemia. Fifty-six different antigens have been serologically defined making the Rh system the most polymorphic of all the erythrocyte blood group systems (1,2). In this article, the molecular bases of the Rh system and the new allele discoveries throughout this time will be reviewed. Also, the impact of the molecular identification of different RH allelic variants in the field of transfusion and obstetric medicine will be discussed.
From antigens to genes
Levine and Stetson (3), in 1939, published the historic case of a mother who required blood transfusion after giving birth a stillborn child and experienced an immediate and severe haemolytic reaction when she was transfused with blood from her husband. Subsequent studies detected an antibody in maternal serum that had an agglutination pattern similar to that obtained with the antibody generated by Landsteiner and Wiener (4) after immunization of rabbits with erythrocytes from Rhesus macaques. Further investigations demonstrated that the specificities of the human and the animal antibodies were different, denoting “anti-Rh” the human antibody and “anti-LW” the animal antibody (5).
According to the theory put forward by Fisher and Race in the 40’s, the Rh system was composed of three close genes, each with two alleles, C and c, D and d, and E and e (6). A person inherited a set of alleles of the three RH genes, known as haplotypes, from each parent. It was originally expected that each allele would be found to determine a corresponding antigen but only C, c, D, E and e had been recognized and it was presumed that d was amorphic. Later in 1951, Wiener proposed that the inheritance of Rh antigens was related to a single gene with multiple alleles (R1, R2, R0, Rz, r, r', r'', ry) (7). Each allele determined an agglutinogen which had multiple factors (Rh0, rh', hr', rh'', hr''). For example, the agglutinogen produced by R1 expresses three factors, Rh0, rh', and hr'' (D, C, and e in Fisher-Race terminology). In 1986, Tippett (8) proposed another model, based on an abundance of serological data, in which Rh antigens are determined by only two RH genes, one encoding D with two different alleles (D and non-D, the latter not coding for a recognizable product), and the other encoding the CcEe antigens with four alleles (ce, Ce, cE and CE).
Colin et al. were the first to report, in 1991, the genetic organization of the RH locus in D-positive and D-negatives individuals (9). Southern blot analyses were key to demonstrating that there are two genes in the RH locus, per haploid genome of D-positive individuals. Later it was confirmed that one of these two genes was absent in the RH locus of D-negative individuals, suggesting that the missing gene encodes the D antigen (10-12). The two RH genes, namely RHD and RHCE, are highly homologous and similarly organized in all D-positive Caucasians. In summary:
- The RH locus is composed of two adjacent homologous structural genes denoted RHD and RHCE (Figure 1);
- Caucasian D-positive individuals have either one or two RHD genes per cell while the D-negative phenotype is mainly caused by the absence of the entire RHD gene;
- The RHD gene encodes the RhD protein that expresses the epitopes of the D antigen;
- The RHCE gene has 4 most common allelic forms: RHCE*ce, RHCE*Ce, RHCE*cE and RHCE*CE and each allele determines the expression of two antigens in ce, Ce, cE or CE combination carried by the RhCE protein (RHCE is the collective name of the 4 alleles).

Rh proteins
The RH genes (RHD and RHCE) encode two 417 amino acids Rh polypeptides (RhD and RhCE, respectively) but the initiating methionine residues are post translationally cleaved (13,14). The mature 416 Rh proteins are palmitoylated but not glycosylated and cross the erythrocyte membrane 12 times. The N-terminal and C-terminal amino acid residues reside on the cytoplasmic side and both proteins are predicted to have 6 extracellular loops. They are erythrocyte specific and only expressed on the red blood cell (RBC) membrane if the Rh associated glycoprotein (RhAG) is also present. Rh and RhAG together with the accessory proteins LW, glycophorin B and CD47 are assembled in the “Rh complex” (15). The core of the complex is predicted to be a heterotrimer involving RhD, RhCE and RhAG, stabilized by interactions between the N-terminal and C-terminal domains to which the accessory polypeptides are linked by non-covalent bonds (16). The Rh complex is associated with the Band3/glycophorin. A complex giving rise to a macromolecular structure that seems to participate in gas and cation transport across the membrane. Particularly, RhAG is involved in NH4+ and CO2 exchange (16,17). This macrocomplex also interacts with the spectrin-based membrane skeleton through ankyrin and protein 4.2 and participates in the maintenance of the shape and mechanical properties of the RBC (18-21). As evidence, Rhnull individuals (lacking all Rh antigens) have a mild clinical condition, called Rh-deficiency syndrome, characterized by membrane abnormalities and some degree of haemolytic anemia. Typical hematological features include the presence of stomatocytes and some spherocytes in blood smears, reduced survival of autologous RBCs and increased erythrocyte osmotic fragility (22).
RhBG and RhCG are human RhAG homologues that are mainly expressed in the kidney and thus the “Rh family” currently consists of five proteins—RhD, RhCE, RhAG, RhBG and RhCG (23-25). Contrary to the three other members of the Rh family, RhD and RhCE wouldn’t be a channel for ammonia and might be implicated only in CO2 transport in RBCs but are functionally redundant (26,27).
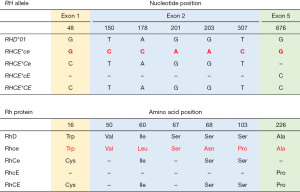
The RhD protein carries the D antigen. The presence or absence of the D antigen in the RBC membrane allows individuals to be classified as “D-positive” or “D-negative”, respectively. The RhCE polypeptide bears the antithetical antigens C or c (involving the second extracellular loop) in addition to E or e (involving the fourth extracellular loop). RhD differs from RhCE by approximately 35 amino acids (depending on the RhCE isoform considered). In spite of this high homology between them, normal RhCE polypeptides do not express any D epitopes and the conventional RhD protein does not express CE antigens (28-30), clearly showing that Rh antigens are conformation-dependent structures. RhCE polypeptides expressing C and c antigens differ in 4 amino acid substitutions—p.Cys16Trp, p.Ile60Leu, p.Ser68Asn and p.Ser103Pro—of which, position 103 is critical for C/c specificity, whereas RhCE polypeptides expressing E and e antigens differ in 1 amino acid change—p.Pro226Ala (31-33) (Figure 2).
RH locus
The RH locus (Figure 1) is located on the short arm of chromosome 1 (position 1p34.1–1p36) (34) and comprises RHD and RHCE. Both genes consist of 10 exons encompassing approximately 58 kb of DNA, reside in tandem in opposite orientation facing each other with their 3' ends, with RHD telomeric of RHCE (9-14,35-37). The region between both genes encompasses 31.8 kb and contains the TMEM50A gene (previously SMP1) (36). Each exon is shorter than 200 bp. RHD and RHCE share a high level of sequence homology (overall 93.8% gene sequence identity and 96.4% exon sequence identity), supporting the concept that these genes have evolved by duplication of a common ancestor (10-14,35,36). Despite their being very closely related, the immunologic heterogeneity within the resulting protein is remarkable. Two 9 kb regions of high homology (98.6%) flank the RHD gene, the so-called 5' Rhesus box and 3' Rhesus box. RHD deletion, the most frequent genetic background responsible for the D-negative phenotype in Caucasians, appears to have occurred through unequal crossing over involving the upstream and downstream Rhesus boxes and the generation of a hybrid Rhesus box in the deleted region. The analysis of the hybrid Rhesus box (a marker for RHD deletion) may be a predictor of the RHD zygosity (36,38).
The coding sequence of RHD (accession number NG_007494.1) contains 37 specific nucleotides that are not found in the 4 most common RHCE alleles (accession number NG_009208.3). RHCE*c has 5 specific nucleotides in exon 2 (c.150C, c.178C, c.201A, c.203A and c.307C), of which the c.307C encoding p.103Pro is best correlated with c expression (31,39). On the other hand, exon 2 of RHCE*C is equal to exon 2 of RHD and only c.48C in exon 1 encoding p.16Cys is RHCE*C specific. However, there is not a strict correlation between c.48C (p.16Cys) and C expression since, mostly in Africans, some RHCE*c alleles with normal expression of c also harbour c.48C in exon 1 (40). Hence RHCE*C genotyping assays must rely on the detection of a 109 bp insertion that is present in intron 2 of RHCE*C alleles only (41). RHCE*E differs from RHCE*e in one single nucleotide variation (SNV) at position 676 in exon 5 (c.676C>G) that leads to the p.Pro226Ala substitution in RhCE proteins expressing E or e respectively. The c.676G is also present in the conventional RHD (31) (Figure 2).
The 3' untranslated region of RHD covers more than 1,500 bp while a shorter stretch is known for RHCE. A deletion of approximately 600 bp in RHD intron 4 is another remarkable difference between RHD and RHCE. Other intronic insertions or deletions over 100 bp have been detected (12,35,42,43). Short tandem repeats are also present in some introns of the RH genes and can be used for polymorphism analysis (35,44,45). GATA-1, SP1 and Ets binding sites are found in the 5' flanking region of RHCE (32).
The discovery of RH alleles over time
Since the cloning and sequencing of the RH genes in the early 1990s and the development of many molecular techniques for the analysis of genetic polymorphisms, a plethora of allelic variants has been identified over time. RH alleles show substantial ethnic variability. Some variants are confined to specific ethnic groups whereas others are more dispersed.
An updated record of RH alleles is available in the International Society of Blood Transfusion (ISBT) website (46). At present (March 2023), 446 RHD alleles and 188 RHCE alleles have been recognized by the ISBT. RHD variants are also listed in the Rhesus Site (47) and many more alleles can be found in published articles and conference abstracts and in databases such as Genbank (48), Erythrogene (49) and RHeference (50).
RH alleles have arisen through different mechanisms such as rearrangements between the RHD and RHCE genes by unidirectional segmental DNA exchanges. This gene conversion event, favoured for the high homology and opposite orientation of both genes, is responsible for the generation of hybrid RH alleles. Many kilobases, including multiple exons and introns can be converted, but often microconversion events, involving a few bases, produce templated mutations (51). In addition, SNVs are responsible for untemplated mutations, where the modified nucleotides do not come from the other gene. Insertions, deletions and duplications are also responsible for the genetic polymorphism of the Rh system. Nucleotide modifications may affect exonic, splice site, intronic and/or promoter regions and lead to amino acid(s) change(s) with subsequent loss or alteration of some epitopes and/or expression of a low incidence antigens or cause premature stop codons preventing protein synthesis. Base changes may also produce frameshifts and splicing alterations leading to shorter or longer polypeptides that will most likely not be integrated in the erythrocyte membrane (46,47,50).
RHD alleles
The RHD*01 allele is the reference sequence for the RHD gene and is responsible for the normal D-positive phenotype. RHD*01.01, characterized by a c.48G>C change, is also considered as to express normal D antigen while the other RHD allelic variants are responsible for D-negative or D variant phenotypes. In this work, we will use the term “D variant” to refer to altered expression of D, such as weak D, partial D or DEL. D variants are serologically recognized because the strength of reactivity with anti-D reagents may be weaker than that of normal D-positive RBCs (≤2+ in tube immediate spin or detected in antiglobulin phase or through adsorption-elution tests) or similar to normal D-positive cells but with the concomitant presence of an anti-D alloantibody. Variants of D can also be recognized when discrepant results using different anti-D reagents are obtained (52).
RHD alleles have been classified according to the encoded phenotype in silent, partial D, weak D and DEL alleles (46,47).
Silent alleles
RHD silent alleles are responsible for a D-negative phenotype. Even though homozygosity for a whole deletion of the RHD gene (RHD*01N.01) is the primary background for the D-negative phenotype in most populations, silent (non-functional) RHD alleles exist that do not generate a complete RhD polypeptide, or the proteins encoded do not express any D epitopes (53-56).
Two RHD silent alleles are frequent in Africans: RHD*08N.01 (RHD*ψ) and RHD*03N.01 (part of r'S haplotype) and at least one copy is harboured by 67% and 15% of D-negative black Africans, respectively. RHD*08N.01 is characterized by a duplication of 37 bp located in the intron 3–exon 4 boundary and 5 nucleotide changes along exons 4 (c.609G>A), exon 5 (c.654G>C, c.667T>G, c.674C>T) and exon 6 (807T>G). This allele may be inactivated by the introduction of a reading frame shift and a translation stop codon at position 210 or by the presence of the nonsense c.807T>G substitution in exon 6 that lead to the p.Tyr269Ter change (57). RHD*08N.01 is generally linked to RHCE*ce.16 that harbours a c.48G>C change in exon 1 and a RHCE-to-RHD gene conversion in exon 9. RHCE*ce.16 has been associated to altered e antigen expression (58). RHD*03N.01 is a hybrid allele in which exons 4 to 8 are derived from RHCE and exon 3 is an RHD-RHCE hybrid but may be completely RHCE (RHD*01N.06). RHD*03N.01 is part of the r'S haplotype together with RHCE*ceS (59-61), characterized by the c.48G>C, c.733C>G, c.1006G>T changes (see RHCE alleles below). In Caucasians, the hybrid RHD-CE(2-9)-D (RHD*01N.03) is the most frequent silent allele encountered and is associated with a R1 haplotype. It is estimated that one copy may be harboured by 0.05% to 0.15% of D-negative individuals (54,55). It is worth mentioning that the African RHD*08N.01 and RHD*03N.01 alleles are also found in whites (56).
There are many other silent RHD alleles responsible for a D-negative phenotype generally harbouring inactivating mutations, such as nucleotide changes, insertions or deletion that give rise to premature stop codons or splice site changes (46,47). These variants are relatively unusual, and the most frequently D-negative genotypes found are homozygosity or compound heterozygosity for RHD deletion, RHD*08N.01, RHD*03N.01 or RHD*01N.03. To note, RHD*01N.75, characterized by a G insertion between positions 581 and 582 (c.581_582insG) in exon 4, is the second null variant most frequently found in D-negative, C- and/or E-positive Argentines, after RHD*03N.01 (62).
RHD silent alleles confound D genotyping because they may lead to false positive results. Molecular strategies must be properly designed considering the distribution of the most frequent silent variants in any given population. Therefore, stringent protocols must be designed to obtain reliable results when D genotyping is performed.
Weak D alleles
Weak D alleles are responsible for a D variant phenotype characterized by a reduced expression of the D antigen. They are most often associated with single nucleotide changes in RHD that lead to RhD proteins with amino acid substitutions predicted to be located in the transmembranous or intracellular segments. Such mutations may negatively affect insertion of proteins in the membrane resulting in reduced amount of D sites at the cell surface (63,64).
Weak D alleles are classified into different “types” according to the mutation responsible for the decreased expression of the D antigen. Some weak D alleles are further divided into several subtypes, for example, weak D type 1.1, weak D type 2.1, weak D type 4.0, weak D type 4.1, etc. The number of weak D alleles is currently greater than 160. However, weak D type 1, weak D type 2, weak D type 3 and weak D type 4 are the most frequently alleles found in D variant individuals, reaching up to 95% in certain populations. DNA-based genotyping protocols have been described to identify the most common weak D alleles in D variant individuals (65-67).
Partial D alleles
Partial D alleles encode RhD proteins with amino acid substitutions predicted to be located in exofacial loops that cause the loss of one or more D specific epitopes (63,64). Partial D individuals can produce anti-D alloantibodies against the absent epitopes following immunization with D-positive RBCs after transfusion or pregnancy. Partial D phenotypes can react weakly with commercial anti-D reagents (for example, the DFR phenotype encoded by RHD*17 alleles), they can also show a strength of hemagglutination equivalent to that observed in D-positive RBCs (for example, the DBT phenotype encoded by RHD*14 alleles) or, even, show an overexpression of the D antigen (for example, the DIVa phenotype encoded by RHD*04 alleles) (1). RHD*06 alleles responsible for the DVI phenotype have been involved in many anti-D alloimmunization events with severe consequences (47,68-70). This has given rise to the development of IgM monoclonal anti-D typing reagents that do not recognize the partial DVI phenotype, the most frequently partial D found in Caucasians. Using this strategy, hemizygous (or the rare homozygous) RHD*06 carriers are typed D-negative and managed accordingly (71,72).
Partial D alleles are mainly generated by DNA segment exchanges between RHD and RHCE. These gene conversion events give rise to hybrid RHD-CE-D or RHCE-D-CE alleles that encode chimeric proteins in which not only some D specific epitopes are missed but also low incidence antigens can be expressed, or high incidence antigens may be lost. Other molecular events such as missense SNVs in one or multiple positions can also lead to partial D alleles (1,46,47).
As mentioned before, monoclonal anti-D reagents cannot reliably distinguish some partial D and weak D phenotypes as they show similar reactivity. DNA-based testing can overcome these limitations and clearly discriminate weak D from partial D phenotypes. The molecular characterization of D variants is useful to implement an appropriate use of D-negative units and a rational administration of anti-D immunoprophylaxis.
DEL alleles
DEL alleles encode mutated RhD proteins that express very low levels of D antigen. The so called DEL phenotype is not detected by standard serology even when D typing is performed with a sensitive indirect antiglobulin test. Therefore, DEL RBCs are frequently mistyped as D-negative unless adsorption-elution tests are performed. The DEL phenotype is almost exclusively found in “seemingly D-negative” individuals expressing the C and/or E antigens (1).
DEL alleles are mainly generated by SNVs in exons or splice sites leading to amino acid changes or affecting RNA splicing. Deletions, insertions, intron polymorphism and hybrid structures have also been described to be responsible for DEL alleles (46,47).
In eastern Asia, between 10% and 33% of individuals who type as D-negative in routine testing are DEL and the most common RHD allele responsible is RHD*DEL1, the so called Asian-type DEL allele (1). It is characterized by a synonymous mutation in the last nucleotide of exon 9 (c.1227G>A, p.Lys409=), that interferes with efficient splicing and results in skipping of exon 9 in the mRNA (73,74).
The incidence of DEL varies from 0.1% to 0.5% among phenotypically D-negative Caucasians and RHD*11 is the most common DEL allele. It carries a c.885G>T substitution in exon 6 leading to p.Met295Ile change in the RhD protein. RHD*11 may occur in two haplotypes, in R0 encodes a weak D phenotype easily demonstrable in the indirect agglutination test but in R1 the antigen density is much lower resulting in a DEL phenotype. The second most common DEL allele in Caucasians is RHD*DEL8 [RHD(IVS3+1G>A)] characterized by the splice site mutation c.486+1G>A and is associated with a R1 haplotype (54-56). To note, RHD*DEL43, characterized by the c.46T>C change in exon 1, is the DEL variant most frequently found in Argentina (56).
DEL alleles may be responsible for genotype-phenotype discrepancies and their putative presence must be taken into account when performing RHD genotyping.
RHCE alleles
The RHCE gene also bears its own heterogeneity, which is responsible for C, c, E and e polymorphisms. The conventional RHCE alleles are designated as RHCE*ce (RHCE*01), RHCE*Ce (RHCE*02), RHCE*cE (RHCE*03), and RHCE*CE (RHCE*04) which encode RhCE proteins coexpressing C or c together with E or e per polypeptide. RHCE*ce is the reference sequence for the RHCE gene. RHCE*c and RHCE*C differ in 6 nucleotide substitutions while the difference between RHCE*e and RHCE*E is a single nucleotide (31) (Figure 2).
Like the RHD gene, numerous allelic variants of RHCE exist and were generated by the same molecular mechanisms that are responsible for RHD alleles (46,47).
RHCE variants are associated with quantitative and qualitative RhCE changes leading to partial, weak, or no expression of the principal antigens, with altered C and e most frequently encountered. As two copies of RHCE are present per genome, partial CcEe antigens are not regularly detected by serologic tests because they are masked by normal antigens expressed on the RhCE proteins encoded in the other chromosome (1). For instance, RBCs expressing an e variant may wrongly be assumed E homozygous if such variant is not detected by routine typing reagents, or a weakly reactive c variant may be missed if RBCs also carry a normal c antigen.
RHCE*CeCW (RHCE*Ce.08.01) encode the low frequency antigen Cw (Rh8) that is found in approximately 2% of Caucasians and 1% of Blacks. This allele is characterized by a c.122A>G variation in exon 1 leading to a p.Gln41Arg in the RhCe polypeptide. Patients carrying this allele type C-positive; however, they can develop anti-C (or anti-Ce) antibodies following transfusion (75). Another important allele responsible for a variant C antigen is RHD*03N.01 (see RHD silent alleles above). It is mainly found in African Blacks and hence in patients with sickle cell disease (SCD) but also occurs in admixed populations. Even though the polypeptide encoded by this allele does not express any D epitopes it carries a partial C antigen that react weakly with polyclonal anti-C but strongly with commercial monoclonal anti-C reagents. RHD*03N.01 is linked to RHCE*ceS (RHCE*ce.20.03) encoding partial c and e (designated eS) antigens. RHD*03N.01 carriers can produce anti-C, anti-e, and/or anti-Ce alloantibodies following transfusion or pregnancy. These complex and multiple specificities are dubbed anti-hrB (59-61,76).
One further aspect to be considered for RHCE variants is the coding of RhCE polypeptides expressing D-like epitopes. Two significant examples are RHCE*ceHAR (RHCE*ce.22.01), found in individuals of German ancestry, and RHCE*ceCF (RHCE*ce.20.06), found in individuals of African ancestry. In RHCE*ceHAR, exon 5 was replaced by the homologous RHD sequence rendering the hybrid RHCE-D(5)-CE allele. RHCE*ceCF is characterized by the templated nucleotide variations c.48G>C in exon 1, c.697C>G and c.733C>G in exon 5, leading to the amino acid changes p.Trp16Cys, p.Gln233Glu and p.Leu245Val in the encoded Rhce protein. These two RHCE alleles are remarkable because, in the absence of RhD protein, individuals carrying RHCE*ceHAR or RHCE*ceCF type D-positive (3+/4+) with some monoclonal anti-D reagents but D-negative with others. Consequently, if wrongly typed D-positive they can become alloimmunized after transfusion of D-positive units (77,78).
RHCE*ce variants are frequent in individuals with African ancestry, and, as an additional complication, are often linked to partial RHD variants. Patients harbouring haplotypes composed of partial RHD and altered RHCE*ce are at risk of developing clinically significant alloantibodies with complex specificities that are difficult to identify serologically. Genotyping and molecular matching will be beneficial for transfusion support in these patients.
Some RHCE variants do not encode E or e and may even not encode C or c. Individuals homozygous or compound heterozygous for these alleles carry the rare Dc-, DCW- and D-- phenotypes and may develop anti-Rh17 (clinically significant antibody against the high prevalence Rh17 antigen) if exposed to conventional RBCs during transfusion or pregnancy. Homozygosity or compound heterozygosity for totally inactivated RHCE alleles in a D-negative person (lack of RHD) gives rise to the Rhnull phenotype (amorph type). It is worth mentioning that inactive RHAG alleles are also responsible for the Rhnull phenotype (regulator type) in homozygotes or compound heterozygotes, even if they carry unaltered RHD and RHCE. Rhnull patients generally develop anti-Rh29 (clinically significant antibody against the high prevalence Rh29 antigen) after transfusion or pregnancy (1,46,47).
Contribution of the molecular study of the Rh system to transfusion and obstetric medicine fields
Hemagglutination is the “gold standard” method for testing Rh antigens. It is a simple and inexpensive technique that has the appropriate specificity and sensitivity for the correct typing of most patients and donors. However, there are some clinical situations in which serological techniques cannot determine the Rh phenotype accurately. When serology turns inapplicable or inconclusive, an alternative approach is to infer the RBC phenotype identifying the RH alleles at the genomic level. Table 1 summarizes the clinical applications of RH genotyping.
Table 1
| Type patients who have been recently transfused |
| Type RBCs coated with immunoglobulin |
| Type fetus to determine risk for HDN or to guide immunoprophylaxis |
| Type pregnant women to identify weak D and partial D phenotypes to determine candidates for Rh immunoglobulin |
| Type blood recipients to identify weak D and partial D phenotypes to avoid use of limited D-negative blood for transfusion |
| Detect RHD alleles that encode a DEL phenotype |
| Type RH alleles for accurate molecular matching in SCD |
| Determine paternal zygosity for RHD |
| Resolve Rh typing discrepancies |
RBCs, red blood cells; HDN, Haemolytic Disease of the Newborn; SCD, sickle cell disease.
Prenatal RH gene genotyping
One of the first clinical applications that arose after the elucidation of the molecular bases associated with the D-positive and D-negative polymorphism was the prenatal prediction of the fetal D phenotype. In the early 1990s it was already possible to determine the fetal D status by analyzing DNA obtained from amniotic fluid or chorionic villi (79). This fetal RHD genotyping strategy has been used for almost 10 years to identify the fetus who is not at risk of HDN (i.e., predicted to be antigen-negative) in D-negative mothers sensitized with anti-D. If the fetus is D-negative, the pregnancy does not require further monitoring other than normal. On the other hand, if the fetus is D-positive, the pregnancy needs more exhaustive monitoring and, sometimes, medical treatment. Strategies have also been developed to predict expression of antigens carried by RhCE or other blood group proteins in fetus at risk of HDN.
Nowadays, non-invasive testing of cell-free fetal DNA (cffDNA) obtained from maternal plasma is the method of choice for determining fetal antigens since it allows avoiding the adverse events associated with amniocentesis or chorionic villus sampling (80,81). Particularly, non-invasive fetal RHD genotyping, based on the analysis of cffDNA in maternal plasma, not only allows an early risk assessment of HDN in pregnancies with anti-D alloantibodies but also has the potential to avoid antenatal anti-D prophylaxis in the 38% to 40% of D-negative pregnant women carrying an RHD-negative fetus (82). In this scenario, it is particularly relevant to take into consideration that the presence of RH allelic variants (mainly RHD silent alleles and DEL alleles) in D-negative mothers (RHD+, D−) may impede the analysis of cffDNA in maternal plasma or inaccurately predict the D status when such alleles are carried by fetuses. The frequency of RHD+, D− individuals has been estimated to vary between 0.2% and 2.1% in different ethnic groups (54-56). Several protocols considering the genetic background of the population under study have been described to reach high sensitivity and specificity in non-invasive fetal RHD detection (greater than 99.5%) (82).
Molecular characterization of D variants
Commercial anti-D serology reagents cannot reliably distinguish partial D and weak D phenotypes and identify which patients are at risk of anti-D development. Anti-D immunization in weak D patients is unusual, but some cases have been well documented in weak D type 11, weak D type 15, weak D type 21, weak D type 41, weak D type 42, weak D type 45 and weak D type 4.2 (also known as DAR) carriers (47,83-90). It has been reported that D variant patients carrying weak D type 1, weak D type 2 or weak D type 3 alleles do not produce anti-D (84). However, a few cases of anti-D in patients with weak D types 1, 2 and 3 phenotypes were described (91-99), being most of them not clinically significant autoantibodies. The consensus among immunohematologists is to consider that weak D type 1, weak D type 2 or weak D type 3 alleles produce a normal D antigen. In 2015, a work group in USA (100) recommended that molecular RHD characterization be implemented in the management of obstetric patients and potential recipients of a blood transfusion with a serologic weak D phenotype, as it was estimated that approximately 80% were weak D type 1, weak D type 2 or weak D type 3 carriers not at risk for alloimmunization and not RhIg candidates. Based on research published since 2015, they updated in 2020 (101) their previous recommendation to include weak D type 4.0 and weak D type 4.1 carriers.
In conclusion, considering the available high-quality evidence from observational studies, it can be assumed that patients with weak D antigen expression caused by weak D type 1, weak D type 2, weak D type 3, weak D type 4.0 or weak D type 4.1 alleles have a very low risk of developing clinically significant anti-D.
Partial D carriers may produce alloantibodies directed to D epitopes missing on their RBCs. Particularly, females of child-bearing potential should receive D-negative RBCs for transfusion and pregnant women must be given RhIg prophylaxis. However, antibody-based typing does not have the capacity to accurately identify partial D phenotypes. Transfusion services often adopt a behaviour aimed at protecting patients from alloimmunization to the D antigen and manage as D-negative all women of child-bearing age with variable or weak D typing. The molecular identification of weak D type 1, weak D type 2, weak D type 3, weak D type 4.0 or weak D type 4.1 carriers in D variant patients may allow managing them safely as D-positive and thus, rationalize the use of D-negative stock units, always in short supply, which could be reserved for patients who are really at risk for anti-D alloimmunization. Moreover, weak D genotyping in D variant pregnant women is a useful tool for guiding the anti-D immunoprophylaxis avoiding unnecessary administration. Molecular characterization of the variant D phenotype also benefits patients who are candidates for chronic transfusions since it would help to decide on adequate transfusion compatibility.
DEL in patients and donors
DEL individuals express a minimum amount of D antigen sites in the RBC membrane (<100 versus 10,000 to 33,000 sites on regular D-positive erythrocytes) (1) and are better diagnosed by RHD genotyping.
Asian-type DEL patients (the DEL phenotype produced by RHD*DEL1) are not prone to produce anti-D. It was reported that no Chinese Asian-type DEL women with a history of pregnancy had developed anti-D, however, this alloantibody was found in truly D-negative pregnant women (102). A recent clinical trial has shown that individuals with Asian-type DEL do not develop anti-D alloantibodies when exposed to D-positive RBCs after transfusion or pregnancy (103). On the contrary, DEL Caucasian individuals with anti-D have been documented, with one of the antibodies causing mild HDN (104,105). It has been suggested that the D antigen that is present in DEL phenotype may have a partial or complete expression. Studies based on adsorption-elution tests with monoclonal anti-Ds reagents have revealed that RHD*DEL1 (the Asian-type DEL allele) encode a RhD protein that express all D epitopes (104). Similar findings have been found with in vitro expression studies (103). In contrast, RHD*DEL8 and RHD*11 (the most frequent Caucasian DEL alleles) lead to partial D antigens, lacking some D epitopes (104).
DEL management should be adapted to needs of the population (106,107). Due to the low frequency of the DEL phenotype among serologically D-negative Caucasian donors, alloimmunization caused by DEL red cell transfusions to D-negative recipients pose a very low risk. However, the RHD genotyping of D-negative donors at first donation is being performed in some blood services to screen for DEL (54,55,108,109).
RHD zygosity
Hemagglutination has limitations to predict RHD zygosity (RHD copy number) in D-positive individuals as it is deduced from haplotype frequency estimates in any given population (1). An accurate determination of RHD homozygous or RHD hemizygous status in fathers is of significance to provide genetic counselling to women alloimmunized with anti-D. RHD zygosity determination is also important in the study of Rh variants. The analysis of the RHD gene copy number can be carried out based on the molecular identification of a hybrid Rhesus box that is present in the RHD deleted chromosome or by quantitative polymerase chain reaction (PCR) strategies to determine RHD gene dosage (36,38,110-112). Both approaches are complementary and require proper controls. It should be considered that zygosity determination can be confounded by altered hybrid Rhesus boxes or by the presence of certain RHD allelic variants. The most accurate approaches for assessing RHD zygosity are likely to be a multiplex ligation-dependent probe amplification (MLPA) assay (113) and a quantitative multiplex polymerase chain reaction of short fluorescent fragments (QMPSF) method (114) that investigate the copy number of all exons of both the RHD and the RHCE genes.
Molecular matching
Patients with SCD are likely to develop clinically significant anti-C, anti-c or anti-e when compatibility tests are based on serology. Moreover, SCD patients sometimes make complex Rh antibodies that are difficult to identify. This problem arises mainly because African black ethnic groups have an increased incidence of allelic variants that encode partial Rh antigens which serologic reactivity is difficult to analyse. Additionally, altered RHCE are commonly linked to altered RHD encoding partial D, so serologically D-positive patients can also make anti-D. RH genotyping is very useful to aid antibody identification and transfusion decision-making in these patients. It has been demonstrated that molecular RH matching is one potential strategy to reduce alloimmunization in SCD patients needing long-term chronic transfusion support and improve red cell use (115-117).
Conclusions
The progress made in the technology of gene targeting has certainly provided invaluable information to expand our knowledge of the Rh system. A plethora of allelic variants has been thoroughly characterized and the association with Rh antigens, phenotypes and haplotypes has been established. Despite this progress, the biological functions of the Rh system are still under study.
A large amount of clinical data has been accumulated and was of value in defining the clinical importance of different RH alleles in the field of transfusion and obstetric medicine.
Classical haemagglutination is still a robust and straightforward technique with a specificity and sensitivity suitable for typing most patients and donors. Considering that DVI is the most common partial D found in Caucasians, anti-D reagents non nonreactive with partial DVI RBCs in direct tests are selected for typing transfusion receptors and pregnant women. Performing only a direct test avoids the risk of sensitization by classifying DVI as D-negative for transfusion and RhIg prophylaxis. On the other hand, detecting DVI and weak D expression is necessary in blood donors and infants of D-negative mothers at risk for D immunization. Anti-D reagents formulated for the antiglobulin phase (generally IgM/IgG blends) are selected to type this cohort.
Nevertheless, there are some clinical situations in which serologic techniques are not appropriate for determining the Rh phenotype accurately. Molecular methods can be applied to achieve a deeper analysis of the Rh system and overcome the limitations of serology. Both methodologies, in conjunction, are undoubtedly useful to prevent incompatibilities, avoid alloimmunization and haemolytic transfusion reactions, diagnose fetal Rh status, guide anti-D immunoprophylaxis, and optimize RBC survival in patients requiring chronic transfusions.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Emilia Sippert, Carine Prisco Arnoni and Maria Rios) for the series “Novel RH Alleles” published in Annals of Blood. The article has undergone external peer review.
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at https://aob.amegroups.com/article/view/10.21037/aob-23-4/coif). The series “Novel RH Alleles” was commissioned by the editorial office without any funding or sponsorship. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Daniels G. Human Blood Groups. 3rd ed. Wiley Blackwell, Oxford UK. 2013.
- Available online: https://www.isbtweb.org/resource/tableofbloodgroupsystems.html
- Levine P, Stetson RE. An unusual case of intragroup agglutination. JAMA 1939;113:126-7. [Crossref] [PubMed]
- Landsteiner K, Wiener AS. An agglutinable factor in human blood recognized by immune sera for rhesus blood. Proc Soc Exp Biol Med 1940;43:223-5. [Crossref]
- Levine P, Celano MJ, Wallace J, et al. A human D-like antibody. Nature 1963;198:596-7. [Crossref] [PubMed]
- Fisher RA, Race RR. Rh gene frequencies in Britain. Nature 1946;157:48. [Crossref] [PubMed]
- Wiener AS. The Rh-Hr blood types: serology, genetics, and nomenclature. Trans N Y Acad Sci 1951;13:199-204. [Crossref] [PubMed]
- Tippett P. A speculative model for the Rh blood groups. Ann Hum Genet 1986;50:241-7. [Crossref] [PubMed]
- Colin Y, Chérif-Zahar B, Le Van Kim C, et al. Genetic basis of the RhD-positive and RhD-negative blood group polymorphism as determined by Southern analysis. Blood 1991;78:2747-52. [Crossref] [PubMed]
- Le van Kim C, Mouro I, Chérif-Zahar B, et al. Molecular cloning and primary structure of the human blood group RhD polypeptide. Proc Natl Acad Sci U S A 1992;89:10925-9. [Crossref] [PubMed]
- Kajii E, Umenishi F, Iwamoto S, et al. Isolation of a new cDNA clone encoding an Rh polypeptide associated with the Rh blood group system. Hum Genet 1993;91:157-62. [Crossref] [PubMed]
- Arce MA, Thompson ES, Wagner S, et al. Molecular cloning of RhD cDNA derived from a gene present in RhD-positive, but not RhD-negative individuals. Blood 1993;82:651-5. [Crossref] [PubMed]
- Chérif-Zahar B, Bloy C, Le Van Kim C, et al. Molecular cloning and protein structure of a human blood group Rh polypeptide. Proc Natl Acad Sci U S A 1990;87:6243-7. [Crossref] [PubMed]
- Avent ND, Ridgwell K, Tanner MJ, et al. cDNA cloning of a 30 kDa erythrocyte membrane protein associated with Rh (Rhesus)-blood-group-antigen expression. Biochem J 1990;271:821-5. [Crossref] [PubMed]
- Hartel-Schenk S, Agre P. Mammalian red cell membrane Rh polypeptides are selectively palmitoylated subunits of a macromolecular complex. J Biol Chem 1992;267:5569-74. [Crossref] [PubMed]
- Gruswitz F, Chaudhary S, Ho JD, et al. Function of human Rh based on structure of RhCG at 2.1 A. Proc Natl Acad Sci U S A 2010;107:9638-43. [Crossref] [PubMed]
- Endeward V, Cartron JP, Ripoche P, et al. RhAG protein of the Rhesus complex is a CO2 channel in the human red cell membrane. FASEB J 2008;22:64-73. [Crossref] [PubMed]
- Nicolas V, Mouro-Chanteloup I, Lopez C, et al. Functional interaction between Rh proteins and the spectrin-based skeleton in erythroid and epithelial cells. Transfus Clin Biol 2006;13:23-8. [Crossref] [PubMed]
- Bruce LJ, Beckmann R, Ribeiro ML, et al. A band 3-based macrocomplex of integral and peripheral proteins in the RBC membrane. Blood 2003;101:4180-8. [Crossref] [PubMed]
- Bruce LJ, Guizouarn H, Burton NM, et al. The monovalent cation leak in overhydrated stomatocytic red blood cells results from amino acid substitutions in the Rh-associated glycoprotein. Blood 2009;113:1350-7. [Crossref] [PubMed]
- Mohandas N, Gallagher PG. Red cell membrane: past, present, and future. Blood 2008;112:3939-48. [Crossref] [PubMed]
- Huang CH. The human Rh50 glycoprotein gene. Structural organization and associated splicing defect resulting in Rh(null) disease. J Biol Chem 1998;273:2207-13. [Crossref] [PubMed]
- Liu Z, Chen Y, Mo R, et al. Characterization of human RhCG and mouse Rhcg as novel nonerythroid Rh glycoprotein homologues predominantly expressed in kidney and testis. J Biol Chem 2000;275:25641-51. [Crossref] [PubMed]
- Liu Z, Peng J, Mo R, et al. Rh type B glycoprotein is a new member of the Rh superfamily and a putative ammonia transporter in mammals. J Biol Chem 2001;276:1424-33. [Crossref] [PubMed]
- Marini AM, Matassi G, Raynal V, et al. The human Rhesus-associated RhAG protein and a kidney homologue promote ammonium transport in yeast. Nat Genet 2000;26:341-4. [Crossref] [PubMed]
- Westhoff CM, Ferreri-Jacobia M, Mak DO, et al. Identification of the erythrocyte Rh blood group glycoprotein as a mammalian ammonium transporter. J Biol Chem 2002;277:12499-502. [Crossref] [PubMed]
- Soupene E, Inwood W, Kustu S. Lack of the Rhesus protein Rh1 impairs growth of the green alga Chlamydomonas reinhardtii at high CO2. Proc Natl Acad Sci U S A 2004;101:7787-92. [Crossref] [PubMed]
- Avent ND, Liu W, Warner KM, et al. Immunochemical analysis of the human erythrocyte Rh polypeptides. J Biol Chem 1996;271:14233-9. [Crossref] [PubMed]
- Huang CH. Molecular insights into the Rh protein family and associated antigens. Curr Opin Hematol 1997;4:94-103. [Crossref] [PubMed]
- Cartron JP, Bailly P, Le Van Kim C, et al. Insights into the structure and function of membrane polypeptides carrying blood group antigens. Vox Sang 1998;74:29-64. [Crossref] [PubMed]
- Mouro I, Colin Y, Chérif-Zahar B, et al. Molecular genetic basis of the human Rhesus blood group system. Nat Genet 1993;5:62-5. [Crossref] [PubMed]
- Chérif-Zahar B, Le Van Kim C, Rouillac C, et al. Organization of the gene (RHCE) encoding the human blood group RhCcEe antigens and characterization of the promoter region. Genomics 1994;19:68-74. [Crossref] [PubMed]
- Smythe JS, Avent ND, Judson PA, et al. Expression of RHD and RHCE gene products using retroviral transduction of K562 cells establishes the molecular basis of Rh blood group antigens. Blood 1996;87:2968-73. [Crossref] [PubMed]
- Chérif-Zahar B, Mattéi MG, Le Van Kim C, et al. Localization of the human Rh blood group gene structure to chromosome region 1p34.3-1p36.1 by in situ hybridization. Hum Genet 1991;86:398-400. [Crossref] [PubMed]
- Okuda H, Suganuma H, Kamesaki T, et al. The analysis of nucleotide substitutions, gaps, and recombination events between RHD and RHCE genes through complete sequencing. Biochem Biophys Res Commun 2000;274:670-83. [Crossref] [PubMed]
- Wagner FF, Flegel WA. RHD gene deletion occurred in the Rhesus box. Blood 2000;95:3662-8. [Crossref] [PubMed]
- Suto Y, Ishikawa Y, Hyodo H, et al. Gene organization and rearrangements at the human Rhesus blood group locus revealed by fiber-FISH analysis. Hum Genet 2000;106:164-71. [Crossref] [PubMed]
- Chiu RW, Murphy MF, Fidler C, et al. Determination of RhD zygosity: comparison of a double amplification refractory mutation system approach and a multiplex real-time quantitative PCR approach. Clin Chem 2001;47:667-72. [Crossref] [PubMed]
- Faas BH, Beuling EA, Ligthart PC, et al. Partial expression of RHc on the RHD polypeptide. Transfusion 2001;41:1136-42. [Crossref] [PubMed]
- Westhoff CM, Silberstein LE, Wylie DE, et al. 16Cys encoded by the RHce gene is associated with altered expression of the e antigen and is frequent in the R0 haplotype. Br J Haematol 2001;113:666-71. [Crossref] [PubMed]
- Tax MG, van der Schoot CE, van Doorn R, et al. RHC and RHc genotyping in different ethnic groups. Transfusion 2002;42:634-44. [Crossref] [PubMed]
- Avent ND, Martin PG, Armstrong-Fisher SS, et al. Evidence of genetic diversity underlying Rh D-, weak D (Du), and partial D phenotypes as determined by multiplex polymerase chain reaction analysis of the RHD gene. Blood 1997;89:2568-77. [Crossref] [PubMed]
- Okuda H, Kawano M, Iwamoto S, et al. The RHD gene is highly detectable in RhD-negative Japanese donors. J Clin Invest 1997;100:373-9. [Crossref] [PubMed]
- Kemp TJ, Poulter M, Carritt B. Microsatellite variation within the human RHCE gene. Vox Sang 1999;77:159-63. [Crossref] [PubMed]
- Fujiwara H, Okuda H, Omi T, et al. The STR polymorphisms in intron 8 may provide information about the molecular evolution of RH haplotypes. Hum Genet 1999;104:301-6. [Crossref] [PubMed]
- Available online: http://www.isbtweb.org/working-parties/red-cell-immunogenetics-and-blood-group-terminology/
- Wagner FF, Flegel WA. The Rhesus Site. Transfus Med Hemother 2014;41:357-63. [Crossref] [PubMed]
- Benson DA, Cavanaugh M, Clark K, et al. GenBank. Nucleic Acids Res 2013;41:D36-42. [Crossref] [PubMed]
- Möller M, Jöud M, Storry JR, et al. Erythrogene: a database for in-depth analysis of the extensive variation in 36 blood group systems in the 1000 Genomes Project. Blood Adv 2016;1:240-9. [Crossref] [PubMed]
- Floch A, Téletchéa S, Tournamille C, et al. A Review of the Literature Organized Into a New Database: RHeference. Transfus Med Rev 2021;35:70-7. [Crossref] [PubMed]
- Fichou Y. Hybrids and microconversions in RH genes: investigation and implication in transfusion therapy. Ann Blood 2023;8:7. [Crossref]
- Daniels G. Variants of RhD--current testing and clinical consequences. Br J Haematol 2013;161:461-70. [Crossref] [PubMed]
- Wagner FF, Frohmajer A, Flegel WA. RHD positive haplotypes in D negative Europeans. BMC Genet 2001;2:10. [Crossref] [PubMed]
- Polin H, Danzer M, Gaszner W, et al. Identification of RHD alleles with the potential of anti-D immunization among seemingly D- blood donors in Upper Austria. Transfusion 2009;49:676-81. [Crossref] [PubMed]
- Flegel WA, von Zabern I, Wagner FF. Six years' experience performing RHD genotyping to confirm D- red blood cell units in Germany for preventing anti-D immunizations. Transfusion 2009;49:465-71. [Crossref] [PubMed]
- Trucco Boggione C, Luján Brajovich ME, Tarragó M, et al. Molecular structures identified in serologically D- samples of an admixed population. Transfusion 2014;54:2456-62. [Crossref] [PubMed]
- Singleton BK, Green CA, Avent ND, et al. The presence of an RHD pseudogene containing a 37 base pair duplication and a nonsense mutation in africans with the Rh D-negative blood group phenotype. Blood 2000;95:12-8. [Crossref] [PubMed]
- Lavoie J, Éthier C, St-Louis M. A new RHCE variant allele, RHCE*48C,1170T,1193A. Transfusion 2016;56:1915-7. [Crossref] [PubMed]
- Faas BH, Beckers EA, Wildoer P, et al. Molecular background of VS and weak C expression in blacks. Transfusion 1997;37:38-44. [Crossref] [PubMed]
- Daniels GL, Faas BH, Green CA, et al. The VS and V blood group polymorphisms in Africans: a serologic and molecular analysis. Transfusion 1998;38:951-8. [Crossref] [PubMed]
- Pham BN, Peyrard T, Juszczak G, et al. Heterogeneous molecular background of the weak C, VS+, hr B-, Hr B- phenotype in black persons. Transfusion 2009;49:495-504. [Crossref] [PubMed]
- Trucco Boggione C, Mufarrege N, Dorigan de Macedo M, et al. Molecular study of D negative and D variant phenotypes in Argentine. Vox Sang 2017;112:abstr 54.
- Wagner FF, Gassner C, Müller TH, et al. Molecular basis of weak D phenotypes. Blood 1999;93:385-93. [Crossref] [PubMed]
- Flegel WA, Denomme GA, Yazer MH. On the complexity of D antigen typing: a handy decision tree in the age of molecular blood group diagnostics. J Obstet Gynaecol Can 2007;29:746-52. [Crossref] [PubMed]
- Gassner C, Schmarda A, Kilga-Nogler S, et al. RHD/CE typing by polymerase chain reaction using sequence-specific primers. Transfusion 1997;37:1020-6. [Crossref] [PubMed]
- Müller TH, Wagner FF, Trockenbacher A, et al. PCR screening for common weak D types shows different distributions in three Central European populations. Transfusion 2001;41:45-52. [Crossref] [PubMed]
- Fichou Y, Le Maréchal C, Jamet D, et al. Establishment of a medium-throughput approach for the genotyping of RHD variants and report of nine novel rare alleles. Transfusion 2013;53:1821-8. [Crossref] [PubMed]
- Mouro I, Le Van Kim C, Rouillac C, et al. Rearrangements of the blood group RhD gene associated with the DVI category phenotype. Blood 1994;83:1129-35. [Crossref] [PubMed]
- Ye L, Wang P, Gao H, et al. Partial D phenotypes and genotypes in the Chinese population. Transfusion 2012;52:241-6. [Crossref] [PubMed]
- von Zabern I, Wagner FF, Moulds JM, et al. D category IV: a group of clinically relevant and phylogenetically diverse partial D. Transfusion 2013;53:2960-73. [Crossref] [PubMed]
- Jones J, Scott ML, Voak D. Monoclonal anti-D specificity and Rh D structure: criteria for selection of monoclonal anti-D reagents for routine typing of patients and donors. Transfus Med 1995;5:171-84. [Crossref] [PubMed]
- Wagner FF, Gassner C, Muller TH, et al. Three molecular structures cause rhesus D category VI phenotypes with distinct immunohematologic features. Blood 1998;91:2157-68. [Crossref] [PubMed]
- Shao CP, Xiong W, Zhou YY. Multiple isoforms excluding normal RhD mRNA detected in Rh blood group Del phenotype with RHD 1227A allele. Transfus Apher Sci 2006;34:145-52. [Crossref] [PubMed]
- Liu HC, Eng HL, Yang YF, et al. Aberrant RNA splicing in RHD 7-9 exons of DEL individuals in Taiwan: a mechanism study. Biochim Biophys Acta 2010;1800:565-73. [Crossref] [PubMed]
- Mouro I, Colin Y, Sistonen P, et al. Molecular basis of the RhCW (Rh8) and RhCX (Rh9) blood group specificities. Blood 1995;86:1196-201. [Crossref] [PubMed]
- Westhoff CM, Vege S, Halter-Hipsky C, et al. DIIIa and DIII Type 5 are encoded by the same allele and are associated with altered RHCE*ce alleles: clinical implications. Transfusion 2010;50:1303-11. [Crossref] [PubMed]
- Beckers EA, Faas BH, von dem Borne AE, et al. The R0Har RH:33 phenotype results from substitution of exon 5 of the RHCE gene by the corresponding exon of the RHD gene. Br J Haematol 1996;92:751-7. [Crossref] [PubMed]
- Flegel WA, Wagner FF, Chen Q, et al. The RHCE allele ceCF: the molecular basis of Crawford (RH43). Transfusion 2006;46:1334-42. [Crossref] [PubMed]
- Bennett PR, Le Van Kim C, Colin Y, et al. Prenatal determination of fetal RhD type by DNA amplification. N Engl J Med 1993;329:607-10. [Crossref] [PubMed]
- Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet 1997;350:485-7. [Crossref] [PubMed]
- Lo YM, Tein MS, Lau TK, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet 1998;62:768-75. [Crossref] [PubMed]
- Ellen van der Schoot C, Winkelhorst D, Clausen FB. Chapter 8 - Noninvasive Fetal Blood Group Typing. In: editors: Page-Christiaens L, Klein HG. Noninvasive Prenatal Testing (NIPT): Applied Genomics in Prenatal Screening and Diagnosis. Academic Press; 2018:125-56.
- Hemker MB, Ligthart PC, Berger L, et al. DAR, a new RhD variant involving exons 4, 5, and 7, often in linkage with ceAR, a new Rhce variant frequently found in African blacks. Blood 1999;94:4337-42. [Crossref] [PubMed]
- Wagner FF, Frohmajer A, Ladewig B, et al. Weak D alleles express distinct phenotypes. Blood 2000;95:2699-708. [Crossref] [PubMed]
- Martinez Badas M, Garcia Sanchez F, Moreno Jimenez G, et al. P-355: Anti-D immunization in a D-positive mother. Vox Sang 2007;93:187.
- Flegel WA, von Zabern I, Doescher A, et al. D variants at the RhD vestibule in the weak D type 4 and Eurasian D clusters. Transfusion 2009;49:1059-69. [Crossref] [PubMed]
- McGann H, Wenk RE. Alloimmunization to the D antigen by a patient with weak D type 21. Immunohematology 2010;26:27-9. [Crossref] [PubMed]
- St-Louis M, Richard M, Côté M, et al. Weak D type 42 cases found in individuals of European descent. Immunohematology 2011;27:20-4. [Crossref] [PubMed]
- Pham BN, Roussel M, Gien D, et al. Molecular analysis of patients with weak D and serologic analysis of those with anti-D (excluding type 1 and type 2). Immunohematology 2013;29:55-62. [Crossref] [PubMed]
- Srivastava K, Polin H, Sheldon SL, et al. The DAU cluster: a comparative analysis of 18 RHD alleles, some forming partial D antigens. Transfusion 2016;56:2520-31. [Crossref] [PubMed]
- Beckers E, Ligthart P, Overbeeke M, et al. A patient with weak D type 1 and anti-D: auto or allo? Vox Sang 2004;87:abstr 75.
- Roxby D, Coloma M, Flegel W, et al. Observation of anti-D after D-positive transfusion in an individual with weak D type-1 phenotype. Vox Sang 2004;87:S77-8.
- Daniels G, Poole G, Poole J. Partial D and weak D: can they be distinguished? Transfus Med 2007;17:145-6. [Crossref] [PubMed]
- Pelc-Klopotowska M, Guz K, Walaszcyk A, et al. Weak D type 2 patient with allo anti-D. Vox Sang 2010;99:abstr 367.
- Vege S, Fong C, Lomas-Francis C, et al. Weak D Type 2 and Production of Anti-D. Transfusion 2011;51:41A.
- Pham BN, Roussel M, Peyrard T, et al. Anti-D investigations in individuals expressing weak D Type 1 or weak D Type 2: allo- or autoantibodies? Transfusion 2011;51:2679-85. [Crossref] [PubMed]
- Nixon C, Ochoa-Garay G, Sweeney J. A Patient with Weak D Type 3 and Anti-D Alloimmunization. Transfusion 2016;56:33A.
- Woo JS, Gikas A, Moayeri M, et al. IGT14-ST4-23: Robust allo-anti-D with subsequent anti-K production after transfusion of D-positive RBCs to a patient with weak D type 1. Transfusion 2018;58:42A.
- Castilho L, Delfino dos Santos T, Menegati S, et al. RHD*weak D type 3 and production of allo-anti-D in a patient with sickle cell disease (SCD). Vox Sang 2019;114:190.
- Sandler SG, Flegel WA, Westhoff CM, et al. It’s time to phase in RHD genotyping for patients with a serologic weak D phenotype. College of American Pathologists Transfusion Medicine Resource Committee Work Group. Transfusion 2015;55:680-9. [Crossref] [PubMed]
- Flegel WA, Denomme GA, Queenan JT, et al. It's time to phase out "serologic weak D phenotype" and resolve D types with RHD genotyping including weak D type 4. Transfusion 2020;60:855-9. [Crossref] [PubMed]
- Shao CP. Transfusion of RhD-positive blood in "Asia type" DEL recipients. N Engl J Med 2010;362:472-3. Erratum in: N Engl J Med 2010;362:1056. [Crossref] [PubMed]
- Ji Y, Luo Y, Wen J, et al. Patients with Asian-type DEL can safely be transfused with RhD-positive blood. Blood 2023;141:2141-50. [Crossref] [PubMed]
- Körmöczi GF, Gassner C, Shao CP, et al. A comprehensive analysis of DEL types: partial DEL individuals are prone to anti-D alloimmunization. Transfusion 2005;45:1561-7. [Crossref] [PubMed]
- Gardener G, Legler T, Hyett J, et al. Anti-D in pregnant women with the RHD(IVS3+1G>A)-associated DEL phenotype. Transfusion 2012;52:2016-9. [Crossref] [PubMed]
- Flegel WA, Wagner FF. DEL. Blood Transfus 2020;18:159-62. [PubMed]
- Ohto H, Flegel WA, Safic Stanic H. When should RhD-negative recipients be spared the transfusion of DEL red cells to avoid anti-D alloimmunization? Transfusion 2022;62:2405-8. [Crossref] [PubMed]
- Crottet SL, Henny C, Meyer S, et al. Implementation of a mandatory donor RHD screening in Switzerland. Transfus Apher Sci 2014;50:169-74. [Crossref] [PubMed]
- Mota M, Dezan M, Valgueiro MC, et al. RHD allelic identification among D-Brazilian blood donors as a routine test using pools of DNA. J Clin Lab Anal 2012;26:104-8. [Crossref] [PubMed]
- Pertl B, Pieber D, Panzitt T, et al. RhD genotyping by quantitative fluorescent polymerase chain reaction: a new approach. BJOG 2000;107:1498-502. [Crossref] [PubMed]
- Chiu RW, Murphy MF, Fidler C, et al. Technical optimization of RhD zygosity determination by real-time quantitative polymerase chain reaction: implication for fetal RhD status determination by maternal plasma. Ann N Y Acad Sci 2001;945:156-60. [Crossref] [PubMed]
- Krog GR, Clausen FB, Dziegiel MH. Quantitation of RHD by real-time polymerase chain reaction for determination of RHD zygosity and RHD mosaicism/chimerism: an evaluation of four quantitative methods. Transfusion 2007;47:715-22. [Crossref] [PubMed]
- Haer-Wigman L, Veldhuisen B, Jonkers R, et al. RHD and RHCE variant and zygosity genotyping via multiplex ligation-dependent probe amplification. Transfusion 2013;53:1559-74. [Crossref] [PubMed]
- Fichou Y, Le Maréchal C, Bryckaert L, et al. A convenient qualitative and quantitative method to investigate RHD-RHCE hybrid genes. Transfusion 2013;53:2974-82. [Crossref] [PubMed]
- Chou ST, Jackson T, Vege S, et al. High prevalence of red blood cell alloimmunization in sickle cell disease despite transfusion from Rh-matched minority donors. Blood 2013;122:1062-71. [Crossref] [PubMed]
- Wilkinson K, Harris S, Gaur P, et al. Molecular blood typing augments serologic testing and allows for enhanced matching of red blood cells for transfusion in patients with sickle cell disease. Transfusion 2012;52:381-8. [Crossref] [PubMed]
- Chou ST, Evans P, Vege S, et al. RH genotype matching for transfusion support in sickle cell disease. Blood 2018;132:1198-207. [Crossref] [PubMed]
Cite this article as: Cotorruelo C. Elucidation of the molecular bases of the Rh system and its contribution to transfusion and obstetric medicine—historical and current perspective: a review. Ann Blood 2023;8:37.


