
Now in 3D! Novel insights into CD36 structure and function
Introduction
CD36 is a cell surface receptor with wide cellular distribution. Its affinity for diverse ligands and distinctive structural features enable it to perform an impressive array of biological functions. Among the many cell types expressing CD36 are adipocytes (1), myocytes (2), monocytes/macrophages (3), microvascular endothelial cells (4), and platelets (5). In adipocytes and myocytes, CD36 (also known as fatty acid translocase) contributes to approximately 50% of total long chain fatty acid uptake (1). Additionally, CD36-mediated fatty acid uptake can be harnessed by tumor cells as a means to accelerate their own growth (6). CD36 present on sensory epithelial cells of the tongue mediates preference for lipid-rich foods (7), while CD36 in the small intestinal epithelium assists in fat-induced satiety (8). In macrophages, CD36 binds oxidized low density lipoprotein (oxLDL), and the excess uptake of oxLDL-associated cholesterol leads to the formation of lipid-laden macrophage foam cells that eventually progress to atherosclerotic lesions (9). Macrophage CD36 also assists in the recognition, phagocytosis, and clearance of apoptotic cells upon oxidized phosphatidylserine exposure (10), a process that is critical for resolving inflammation (11). Endothelial cell CD36 binds the matricellular glycoprotein thrombospondin-1, which regulates angiogenesis (12), and also tethers Plasmodium falciparum (P. falciparum)-infected erythrocytes to the vascular wall, thus preventing their splenic clearance and prolonging malarial infection (13-15). In platelets, binding of CD36 to oxLDL and other danger-associated molecular pattern structures promotes platelet activation and a pro-thrombotic state associated with chronic inflammatory disorders (16-18). For an excellent review of this subject, see Yang et al. (19). Additionally, CD36 (also known as platelet glycoprotein IV) binds the adhesive glycoprotein thrombospondin-1, which promotes platelet adhesion and thrombus stabilization (20).
The exceptional diversity of CD36-mediated biological functions is made possible, in part, by various structural features and post-translational modifications. Over the last three decades, researchers have employed various biochemical strategies that have led to the identification of several important structural features of CD36. However, due to the lack of a CD36 structure, many of these studies relied upon structural feature prediction software and homology modeling. Recently, a high-resolution extracellular crystal structure of CD36 was determined (21), offering additional insights into CD36 structure-function relationships. In this narrative literature review, structural features relevant to platelet biology will be emphasized. To obtain relevant literature, our search results were limited to publications within the PubMed database using the keywords “CD36” in combination with other words that include “structure”, “platelet”, “mutant”, and “oligomerization”.
General CD36 structural features
CD36 is a 472-amino-acid transmembrane glycoprotein and member of the class B scavenger receptor family, which includes the related proteins lysosomal integral membrane protein-2 (LIMP-2) and scavenger receptor class B type I (SR-BI). Like all class B scavenger receptors, the overall structure of CD36 consists of a large extracellular domain that is anchored to the cell membrane by two transmembrane domains adjacent to short N- and C-terminal cytosolic tails (15,22). The amino acid sequence of human CD36 can be subcategorized into five regions: N-terminal cytosolic domain (residues 1–7), N-terminal transmembrane domain that also functions as a noncleaved signal peptide (residues 8–29), extracellular domain (residues 30–439), C-terminal transmembrane domain (residues 440–461), and C-terminal cytosolic domain (residues 462–472) (Figure 1). While each subdomain plays a unique role in maintaining CD36 function, the extracellular domain is perhaps most critical, as it binds to the various ligands and facilitates fatty acid uptake.
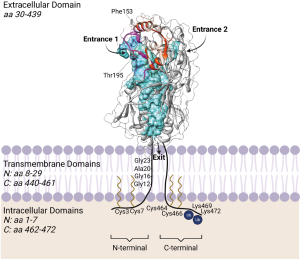
High-resolution structure of CD36
Until recently, structure-function studies of CD36 were hindered by the lack of a high-resolution CD36 structure, and researchers relied on various biochemical techniques (e.g., mutagenesis) to predict biologically-relevant structural features of CD36. However, interpretation of mutagenesis studies can be limited in cases where a mutation unexpectedly reduces protein stability or expression. With the recent structural determination of the extracellular domain of human CD36 (residues 35–439) by X-ray crystallography (21), we now have a detailed blueprint to better understand the mechanisms underlying CD36 function. As predicted by homology modeling (27), the CD36 extracellular domain adopts an oval-shaped structure with an antiparallel β-barrel core (21). The β-barrel core forms a hydrophobic cavity that traverses the length of the extracellular domain (21). During structure determination, the core cavity was identified as having two possible entrances (termed “entrance 1” and “entrance 2”; Figure 1), both of which were crystallized in the presence of long chain fatty acids (i.e., palmitic acid and stearic acid) (21). Interactions between the long chain fatty acids and the central cavity are primarily mediated by hydrophobic effects between the fatty acid tail and hydrophobic residues lining the cavity, as well as hydrogen bonding between the carboxylic acid head group of fatty acids and Thr195 within the hydrophobic cavity of CD36 (21). This is in agreement with a previous study that utilized homology modeling to predict a fatty acid binding pocket on CD36, which contained Thr195 (28). As CD36 was crystallized in the presence of a CD36 binding domain of P. falciparum erythrocyte membrane protein 1 (PfEMP1), Hsieh and colleagues were able to determine that binding of PfEMP1 to CD36 is largely facilitated by Phe153, a CD36 membrane-distal residue located near entrance 1 of the hydrophobic cavity (21). Further, binding of PfEMP1 competes with oxLDL binding, suggesting that oxLDL binds to the same or overlapping sites on CD36 (21).
The CD36 crystal structure also helps to clarify some structure-function-based studies pre-dating CD36 homology modeling and structural determination. A hydrophobic stretch of amino acids in the CD36 extracellular domain (amino acids 186–204) was previously thought to embed into the outer leaflet of the plasma membrane near the transmembrane domains (29). The CD36 crystal structure appears to contradict this hypothesis, as amino acids 186–204 are membrane distal and some residues in this region face the inner hydrophobic cavity (e.g., Thr195) (Figure 1). Another discrepancy between the CD36 crystal structure and previous studies is the lack of phosphorylation at Thr92 and the relationship between Thr92 phosphorylation status and PfEMP1 binding (as described below). The absence of Thr92 phosphorylation on the CD36 crystal structure holds important implications in platelets, which are thought to be constitutively phosphorylated at Thr92 and thus inhibited from binding thrombospondin-1 in a resting (non-activated) state (22).
CD36 ligands and binding sites
As a member of the scavenger receptor superfamily, CD36 (also known as scavenger receptor class B member 3) binds a remarkable assortment of ligands. These include unmodified and modified lipoproteins (30-32), anionic phospholipids (33), long chain (1) and very long chain (34) fatty acids, P. falciparum-infected erythrocytes (15), thrombospondin-1 (35), pro-inflammatory S100A family proteins (including the myeloid-related protein-8/14 (MRP-8/14) heterodimer) (36), advanced glycation end-products (18,37), growth hormone-releasing peptide hexarelin (38), amyloid-β (39), apoptotic cells (40), endothelial cell-derived microvesicles (41), and, to some extent, collagen (42). Different ligands have been shown to bind to CD36 on the same site, overlapping sites, or distant sites. In this review, we will focus on the binding sites of CD36 ligands that are most relevant to platelet biology, namely, oxLDL, thrombospondin-1, and anionic phospholipids.
The binding site for oxLDL was determined by monoclonal antibody inhibition to be a region spanning amino acids 155–183 on human CD36 (25,26,43) (Figure 2). Within this region, which is located near the apex of the extracellular domain (21,27), the positive charges on Lys164 and Lys166 were identified through mutagenesis studies as critical for oxLDL binding (27,44). Also in the apex region, mutation of Leu158 or Leu161 to negatively-charged glutamate disrupted oxLDL binding (27). Directly adjacent to the proposed oxLDL binding site, mutation of Phe153 to alanine also blocked oxLDL binding to CD36 (21). Altogether, these mutational studies suggest that oxLDL binding to CD36 may be mediated by both hydrophobic and ionic interactions. Other ligands thought to share a binding site with oxLDL and/or compete with oxLDL for binding to CD36 are unmodified and oxidatively-modified lipoproteins (30), long chain fatty acids (45), oxidized phosphatidylcholine (44), P. falciparum-infected erythrocytes/PfEMP1 (21,46), apoptotic neutrophils (47), and hexarelin (48).
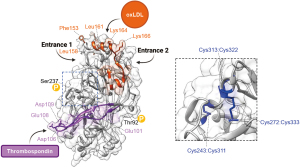
The binding site of thrombospondin-1 is distinct from that of oxLDL (21) and was defined through binding studies of recombinant CD36 peptides (fused to glutathione S-transferase) as a region spanning amino acids 93–120, known as a CD36, LIMP-2, Emp sequence homology (CLESH) domain, which is sufficient for binding thrombospondin-1 type 1 repeats (49,50) (see Figure 2). The binding interface, as revealed by mutagenesis, molecular modeling, and NMR studies, is comprised of several acidic residues on CD36 (Glu101, Asp106, Glu108, and Asp109) that interact with positively-charged residues on thrombospondin (12). As discussed, the binding of thrombospondin-1 to platelet CD36 may be activated by de-phosphorylation of Thr92, which simultaneously reduces collagen binding (22).
Anionic phospholipids (e.g., phosphatidylserine exposed on apoptotic cells, cell-derived microvesicles, or activated platelets) do not share a binding site with thrombospondin or collagen (51). However, phosphatidylserine-containing liposomes were determined in competition assays using different monoclonal antibodies to bind CD36 at a similar but non-identical site to oxLDL, spanning amino acids 162–183 (52).
The large number of CD36 ligands and oftentimes overlapping nature of their binding sites could have important implications for cellular homeostasis. For example, small molecules that inhibit amyloid-β-CD36 interactions were predicted to bind near the exit point (see Figure 1) of the large hydrophobic cavity that shuttles fatty acids into cells (53). Additionally, PfEMP1 was demonstrated to compete with oxLDL for binding to CD36 (21). In these cases, binding of disease-associated ligands (e.g., amyloid-β or PfEMP1) may prevent necessary physiological functions of CD36 (e.g., fatty acid transport).
Post-translational modifications of CD36
To date, several types of post-translational modifications have been identified on CD36, including glycosylation, phosphorylation, ubiquitination, acetylation, and palmitoylation. Many of these modifications play dynamic regulatory roles in CD36 function. While this subject was reviewed in depth by Luiken et al. (54), some of the known CD36 post-translational modifications are briefly summarized below and in Figure 2.
CD36 is heavily glycosylated, which may account for its remarkable resistance to protease digestion, as well as its migration at approximately 88 kDa on SDS-polyacrylamide gel electrophoresis instead of the predicted 53 kDa. The extracellular domain of human CD36 contains 10 putative asparagine (N)-linked glycosylation sites, nine of which are confirmed to be glycosylated (55). Glycosylation of three sites on the carboxy terminal side of CD36 (Asn247, Asn321, and Asn417) is required for trafficking of CD36 to the plasma membrane (55). In macrophages, CD36 was identified as a novel protein partner of sialidase 1 (NEU1), which cleaves terminal sialic acid moieties from glycoproteins (56). CD36 sialylation levels and oxLDL uptake were demonstrated to be regulated by NEU1 activity (56).
Two potential phosphorylation sites have been identified on CD36 at Thr92 and Ser237, which are located within consensus sites for phosphorylation by protein kinase C and protein kinase A, respectively (22,57). However, it is important to note that there is a lack of evidence of CD36 phosphorylation in vivo (58) and in the CD36 crystal structure (21). The exact mechanisms of CD36 phosphorylation are unknown, but there is some evidence that phosphorylation is mediated by ectoprotein kinases (57-59). Phosphorylation status of Thr92 and Ser237 could contribute to the regulation of multiple functions of CD36. Due to its proximity to the CLESH domain, Thr92 phosphorylation inhibits thrombospondin-1 binding to platelets, while it enhances collagen binding (22,58). Interestingly, Thr92 is constitutively phosphorylated on resting platelets, which may explain the lack of thrombospondin-1 binding to resting platelets (22). Others report that Thr92 phosphorylation may also increase the affinity of CD36 for P. falciparum-infected erythrocytes (60). Contradicting these previous studies, the crystal structure of CD36 lacks Thr92 phosphorylation and shows that Thr92 is distal from the binding site of P. falciparum-infected erythrocytes (21) (Figure 2). Thus, it is unlikely that Thr92 phosphorylation blocks PfEMP1 binding. This is not entirely unexpected, as P. falciparum-infected erythrocytes and thrombospondin-1 are thought to bind non-overlapping sites on CD36. Nonetheless, more work is needed to reconcile these conflicting results. Less is known about a potential role for Ser237 phosphorylation in regulating CD36 function, but there is a single report that it inhibits fatty acid uptake in platelets (59). There was no mention of phosphorylation at Ser237 in the recent CD36 crystal structure (21).
CD36 is potentially ubiquitinated at Lys469 and Lys472, which regulates protein stability (61). In C2C12 myotubes and CD36/insulin receptor-overexpressing CHO cells, it was demonstrated that CD36 poly-ubiquitination and subsequent degradation increase with fatty acid treatment and decrease with insulin treatment (61). Mutation of both Lys469 and Lys472 to alanine almost completely blocked CD36 ubiquitination (61). The relative individual contributions of Lys469 and Lys472 to CD36 poly-ubiquitination were not investigated. Various de-ubiquitinases have recently been implicated in removing ubiquitin from CD36, including ubiquitin-specific peptidases 10 and 14 (USP10 and USP14) and ubiquitin C-terminal hydrolase L1 (UCHL1), and inhibition of each of these enzymes led to CD36 degradation and reduced foam cell formation (62-64). While UCHL1 expression is localized primarily to brain tissues, USP10 and USP14 are ubiquitously expressed (65). Therefore, USP10- and USP14-mediated increases in CD36 stability may be particularly relevant to other cell types besides macrophages. In contrast to poly-ubiquitination, CD36 mono-ubiquitination by the E3 ubiquitin ligase Parkin is thought to enhance CD36 stability and cell surface expression levels (66,67). Mono-ubiquitin likely occurs on Lys469 and/or Lys472, but this is pending more detailed investigation.
Acetylation has been detected on the extracellular domain of CD36 at the ε-amino position of four lysine residues: Lys52, Lys166, Lys231, and Lys403 (45,68); however, our current understanding of the functional roles of CD36 lysine acetylation remains limited. In general, lysine acetylation neutralizes the positive charge and increases the hydrophobicity of lysine residue sidechains. As Lys166 is required for oxLDL and oxidized phospholipid binding to CD36 (27,44), it is possible that Lys166 acetylation/deacetylation could alter the affinity of ligands for CD36. Protein lysine acetylation may also influence protein synthesis, stability, localization, and cell signaling. The significance of CD36 lysine acetylation is just beginning to be understood. For example, CD36 acetylation/deacetylation events were recently reported to play a role in INS 832/13 beta cell lipid accumulation and apoptosis signaling (69).
CD36 is known to be palmitoylated at cysteine residues within its N-terminal cytosolic domain (Cys3 and Cys7) and C-terminal cytosolic domain (Cys464 and Cys466) (70). CD36 mutants lacking palmitoylation at one or all of these residues retain their ability to express at the cell surface, but they exhibit shorter half-lives, impaired lipid raft localization, and failure to translocate to the plasma membrane in response to insulin or AMP-kinase stimulation (71,72). It was recently demonstrated that treatment of adipocytes with fatty acids induced CD36 de-palmitoylation and internalization (73). The relative contributions of different cysteine palmitoylation sites to CD36 internalization have not been investigated thus far. It was reported that the N-terminal cytosolic domain, unlike the C-terminal cytosolic domain (74), is dispensable for oxLDL uptake (75). Therefore, it may be suggested that Cys464 and/or Cys466 play a more significant role in regulating CD36 stability.
Cysteine disulfide bonds
The extracellular domain of CD36 contains six cysteine residues that were all reported to reside in disulfide bonds (Cys243−Cys311, Cys272−Cys333, and Cys313−Cys322) (76), which has now been confirmed by the CD36 extracellular domain crystal structure (21) (Figure 2). It has been reported that these disulfide bonds play a role in CD36 processing and ER-to-Golgi transport (77) and in CD36 homo-dimerization (78). Studies using chemical reducing agents showed that CD36 extracellular disulfide bonds were required for adherence of P. falciparum-infected erythrocytes but were not required for oxLDL binding (79,80). Recently, the Cys333-Cys272 disulfide bond pair was reported to act as a molecular switch in response to hydrogen sulfide-mediated disulfide bond cleavage (81). Loss of the Cys333-Cys272 disulfide bond may open another entrance to the central hydrophobic cavity that is inaccessible when the disulfide bond is present, resulting in increased uptake of long chain fatty acids and activation of cell signaling pathways promoting gastric cancer cell metastasis (81). More research is needed to determine the possible implications of a Cys333-Cys272 molecular switch in other biological functions of CD36 (e.g., in atherosclerosis development and platelet activation).
CD36 dimerization
CD36 has been previously reported to form homo-dimers and oligomers, which may influence the ability of CD36 to bind ligands or initiate cell signaling events. Few studies thus far have investigated the structural features necessary for dimerization of CD36. A related class B scavenger receptor, SR-BI, has been reported to dimerize via an N-terminal transmembrane glycine motif, as well as a C-terminal transmembrane leucine zipper motif (82-84). CD36 has a corresponding glycine dimerization motif in its N-terminal transmembrane domain, consisting of residues Gly12, Gly16, Ala20, and Gly23 (85). Analysis of both CD36 transmembrane domain segments by fluorescence resonance energy transfer and TOXCAT transmembrane dimer assays indicated that only the N-terminal transmembrane domain dimerizes and that mutation of any of the four glycine motif residues impaired dimerization (85). A single report showed that intermolecular disulfide bonds covalently link CD36 homo-dimers and multimers in the kidney COS-7 cell line and platelets (78). It was inferred that the extracellular cysteine residues mediate dimerization, as mutating all intracellular cysteines did not disrupt dimerization (78). However, this has not been directly tested, and the CD36 structure confirms that all six extracellular cysteine residues are present in intramolecular disulfide bonds (21).
Many questions remain as to the functional role(s) of CD36 dimerization. While SR-BI was reported to form large multimeric complexes on the cell surface that are required for its membrane retention (84), such a phenomenon has not been reported for CD36. Furthermore, loss of SR-BI dimerization correlated with decreased ability of SR-BI to bind high density lipoprotein (HDL) and mediate HDL-cholesterol delivery into cells (82). So far, there have been no reports that CD36 dimerization is required for ligand binding. In fact, purified monomeric CD36 was shown to be capable of binding oxLDL and acetylated LDL with high affinity (86), and recombinant and synthetic CD36 peptides have been shown to bind to oxLDL and thrombospondin-1 (12,44,49). Conversely, ligand binding may activate CD36 dimerization. For instance, thrombospondin-1 triggered CD36 dimerization in transfected HeLa cells, and dimerization required the C-terminal transmembrane domain and/or C-terminal intracellular domain (87). The possibility of ligand-induced conformational changes that facilitate CD36 dimerization or oligomerization requires future investigation. It remains unknown if thrombospondin-1 triggers CD36 homo-dimerization in platelets or if dimerization is required for CD36 function.
CD36-interacting proteins and downstream signaling events
CD36 can form signaling complexes with other transmembrane proteins, including Toll-like receptors (TLRs) (88,89), sodium-potassium-ATPase (NKA) (90), and tetraspanins and integrins (91-94), that engage with downstream signaling molecules. In macrophages, oxLDL and amyloid-β promote CD36-dependent TLR4-TLR6 activation and downstream NF-κB pro-inflammatory signaling (89). OxLDL binding to CD36 also stimulates the formation of a protein complex with NKA and the Src family kinase Lyn, that promotes oxLDL uptake and foam cell formation in macrophages (90). Complexes consisting of β1 and/or β2 integrins, tetraspanins (CD9 and CD81), and Fc receptor γ-chain (FcRγ) adapter protein lead to activation of Src and Syk kinases and CD36 internalization (95). The structural features of CD36 that mediate interactions with other transmembrane proteins have not been characterized, nor is it well established which protein partners interact directly or indirectly with CD36.
The short C-terminal intracellular region of CD36, consisting of approximately 11 amino acids (residues 462–472), directly interacts with protein tyrosine kinase 2 (also known as focal adhesion kinase), the focal adhesion adaptor protein paxillin, and the Src family kinase Lyn (89). In platelets, CD36 has been shown to interact with Src family kinases, including Fyn, Lyn, and Yes (96). CD36 stimulation generates reactive oxygen species (namely hydrogen peroxide), leading to cysteine sulfenylation and activation of Src family kinases (97). Downstream of Src family kinases, caspases and extracellular signal-regulated kinase 5 (ERK5), mediate phosphatidylserine exposure and subsequent thrombosis in response to oxLDL binding to CD36 (16). The interaction of CD36 with Lyn is mediated by Tyr463 of CD36 and is critical for oxLDL-induced TLR4-TLR6 heterodimerization and NF-κB activation (89). Likewise, mutation of Tyr463 or Cys464 on CD36 blocked lipoteichoic acid (a bacterial cell wall component) internalization and subsequent NF-κB activation, indicating a role for these C-terminal residues in other signaling events, including phagocytic pathways (98). Syk kinase also appears to play a role in platelet signaling responses to oxLDL and thrombospondin-1 (99). However, the exact residues or regions of CD36 that interact with Syk kinase have not been established.
Conclusions
Despite the integral role of CD36 in various physiological and pathological states and its possible implications for human health, until recently, we lacked a comprehensive understanding of the structural features contributing to its multitude of functions. In this review, we have summarized our current knowledge of CD36 structural features and highlighted several areas in which more in-depth studies are needed. For example, additional studies must be performed to address the purpose of CD36 acetylation and to clarify if CD36 phosphorylation occurs in vivo. The structural features mediating CD36 dimerization are understudied, as is the function of CD36 dimerization or multimerization (e.g., increased membrane retention). Most importantly, high-resolution structures of the transmembrane and cytosolic domains will provide new insight into the structural mechanisms that facilitate interactions between CD36 and other protein partners and how these interactions may drive downstream signaling events. Potential therapeutic strategies targeting CD36 are being investigated in clinical trials for cancer, but investigations are still in early stages for other conditions, including Alzheimer’s disease, diabetes, atherosclerosis, and thrombosis. One inherent challenge to developing CD36-based therapeutics lies in its ability to perform many cellular functions, including some that are physiologically necessary. With the emergence of new structural information for CD36, perhaps researchers will now be able to target different functions of CD36 with greater specificity.
Acknowledgments
The authors thank Dr. Roy Silverstein, Darcy Knaack, Hayley Powers, and Gage Stuttgen for critical review of this manuscript.
Funding: This work was supported by HL58012 (DS) and HL138907 (DS) from the National Heart Lung and Blood Institute and AI147500 (DS) from the National Institute of Allergy and Infectious Diseases, both of the National Institutes of Health.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Brian R. Curtis) for the series “Thrombocytopenia Due to Immunization Against CD36” published in Annals of Blood. The article has undergone external peer review.
Peer Review File: Available at https://dx.doi.org/10.21037/aob-21-43
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/aob-21-43). The series “Thrombocytopenia Due to Immunization Against CD36” was commissioned by the editorial office without any funding or sponsorship. DS reports that this work was supported by NIH HL58012, NIH HL138907 and NIH AI147500. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Abumrad NA, el-Maghrabi MR, Amri EZ, et al. Cloning of a rat adipocyte membrane protein implicated in binding or transport of long-chain fatty acids that is induced during preadipocyte differentiation. Homology with human CD36. J Biol Chem 1993;268:17665-8. [Crossref] [PubMed]
- Van Nieuwenhoven FA, Verstijnen CP, Abumrad NA, et al. Putative membrane fatty acid translocase and cytoplasmic fatty acid-binding protein are co-expressed in rat heart and skeletal muscles. Biochem Biophys Res Commun 1995;207:747-52. [Crossref] [PubMed]
- Talle MA, Rao PE, Westberg E, et al. Patterns of antigenic expression on human monocytes as defined by monoclonal antibodies. Cell Immunol 1983;78:83-99. [Crossref] [PubMed]
- Swerlick RA, Lee KH, Wick TM, et al. Human dermal microvascular endothelial but not human umbilical vein endothelial cells express CD36 in vivo and in vitro. J Immunol 1992;148:78-83. [PubMed]
- Rhinehart-Jones T, Greenwalt DE. A detergent-sensitive 113-kDa conformer/complex of CD36 exists on the platelet surface. Arch Biochem Biophys 1996;326:115-8. [Crossref] [PubMed]
- Yang P, Su C, Luo X, et al. Dietary oleic acid-induced CD36 promotes cervical cancer cell growth and metastasis via up-regulation Src/ERK pathway. Cancer Lett 2018;438:76-85. [Crossref] [PubMed]
- Laugerette F, Passilly-Degrace P, Patris B, et al. CD36 involvement in orosensory detection of dietary lipids, spontaneous fat preference, and digestive secretions. J Clin Invest 2005;115:3177-84. [Crossref] [PubMed]
- Schwartz GJ, Fu J, Astarita G, et al. The lipid messenger OEA links dietary fat intake to satiety. Cell Metab 2008;8:281-8. [Crossref] [PubMed]
- Febbraio M, Hajjar DP, Silverstein RL. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J Clin Invest 2001;108:785-91. [Crossref] [PubMed]
- Greenberg ME, Sun M, Zhang R, et al. Oxidized phosphatidylserine-CD36 interactions play an essential role in macrophage-dependent phagocytosis of apoptotic cells. J Exp Med 2006;203:2613-25. [Crossref] [PubMed]
- Kourtzelis I, Hajishengallis G, Chavakis T. Phagocytosis of Apoptotic Cells in Resolution of Inflammation. Front Immunol 2020;11:553. [Crossref] [PubMed]
- Klenotic PA, Page RC, Li W, et al. Molecular basis of antiangiogenic thrombospondin-1 type 1 repeat domain interactions with CD36. Arterioscler Thromb Vasc Biol 2013;33:1655-62. [Crossref] [PubMed]
- Barnwell JW, Asch AS, Nachman RL, et al. A human 88-kD membrane glycoprotein (CD36) functions in vitro as a receptor for a cytoadherence ligand on Plasmodium falciparum-infected erythrocytes. J Clin Invest 1989;84:765-72. [Crossref] [PubMed]
- Ockenhouse CF, Tandon NN, Magowan C, et al. Identification of a platelet membrane glycoprotein as a falciparum malaria sequestration receptor. Science 1989;243:1469-71. [Crossref] [PubMed]
- Oquendo P, Hundt E, Lawler J, et al. CD36 directly mediates cytoadherence of Plasmodium falciparum parasitized erythrocytes. Cell 1989;58:95-101. [Crossref] [PubMed]
- Yang M, Kholmukhamedov A, Schulte ML, et al. Platelet CD36 signaling through ERK5 promotes caspase-dependent procoagulant activity and fibrin deposition in vivo. Blood Adv 2018;2:2848-61. [Crossref] [PubMed]
- Podrez EA, Byzova TV, Febbraio M, et al. Platelet CD36 links hyperlipidemia, oxidant stress and a prothrombotic phenotype. Nat Med 2007;13:1086-95. [Crossref] [PubMed]
- Zhu W, Li W, Silverstein RL. Advanced glycation end products induce a prothrombotic phenotype in mice via interaction with platelet CD36. Blood 2012;119:6136-44. [Crossref] [PubMed]
- Yang M, Silverstein RL. CD36 signaling in vascular redox stress. Free Radic Biol Med 2019;136:159-71. [Crossref] [PubMed]
- Kuijpers MJ, van der Meijden PE, Feijge MA, et al. Factor XII regulates the pathological process of thrombus formation on ruptured plaques. Arterioscler Thromb Vasc Biol 2014;34:1674-80. [Crossref] [PubMed]
- Hsieh FL, Turner L, Bolla JR, et al. The structural basis for CD36 binding by the malaria parasite. Nat Commun 2016;7:12837. [Crossref] [PubMed]
- Asch AS, Liu I, Briccetti FM, et al. Analysis of CD36 binding domains: ligand specificity controlled by dephosphorylation of an ectodomain. Science 1993;262:1436-40. [Crossref] [PubMed]
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 2004;25:1605-12. [Crossref] [PubMed]
- Tian W, Chen C, Lei X, et al. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res 2018;46:W363-7. [Crossref] [PubMed]
- Daviet L, Buckland R, Puente Navazo MD, et al. Identification of an immunodominant functional domain on human CD36 antigen using human-mouse chimaeric proteins and homologue-replacement mutagenesis. Biochem J 1995;305:221-4. [Crossref] [PubMed]
- Daviet L, Morel-Kopp MC, Kaplan C, et al. A structural/functional domain on human CD36 is involved in the binding of anti-Nak(a) antibodies. Thromb Haemost 1995;73:543-5. [Crossref] [PubMed]
- Neculai D, Schwake M, Ravichandran M, et al. Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36. Nature 2013;504:172-6. [Crossref] [PubMed]
- Tarhda Z, Semlali O, Kettani A, et al. Three Dimensional Structure Prediction of Fatty Acid Binding Site on Human Transmembrane Receptor CD36. Bioinform Biol Insights 2013;7:369-73. [Crossref] [PubMed]
- Coburn CT, Abumrad NA. Structure-Function of CD36 and Evidence for Its Role in Facilitating Membrane Fatty Acid Transport. In: Duttaroy AK, Spener F. editors. Cellular Proteins and Their Fatty Acids in Health and Disease: Wiley-VCH, 2003:3-30.
- Calvo D, Gómez-Coronado D, Suárez Y, et al. Human CD36 is a high affinity receptor for the native lipoproteins HDL, LDL, and VLDL. J Lipid Res 1998;39:777-88. [Crossref] [PubMed]
- Endemann G, Stanton LW, Madden KS, et al. CD36 is a receptor for oxidized low density lipoprotein. J Biol Chem 1993;268:11811-6. [Crossref] [PubMed]
- Acton SL, Scherer PE, Lodish HF, et al. Expression cloning of SR-BI, a CD36-related class B scavenger receptor. J Biol Chem 1994;269:21003-9. [Crossref] [PubMed]
- Rigotti A, Acton SL, Krieger M. The class B scavenger receptors SR-BI and CD36 are receptors for anionic phospholipids. J Biol Chem 1995;270:16221-4. [Crossref] [PubMed]
- Drover VA, Nguyen DV, Bastie CC, et al. CD36 mediates both cellular uptake of very long chain fatty acids and their intestinal absorption in mice. J Biol Chem 2008;283:13108-15. [Crossref] [PubMed]
- Asch AS, Barnwell J, Silverstein RL, et al. Isolation of the thrombospondin membrane receptor. J Clin Invest 1987;79:1054-61. [Crossref] [PubMed]
- Kerkhoff C, Sorg C, Tandon NN, et al. Interaction of S100A8/S100A9-arachidonic acid complexes with the scavenger receptor CD36 may facilitate fatty acid uptake by endothelial cells. Biochemistry 2001;40:241-8. [Crossref] [PubMed]
- Ohgami N, Nagai R, Ikemoto M, et al. CD36, serves as a receptor for advanced glycation endproducts (AGE). J Diabetes Complications 2002;16:56-9. [Crossref] [PubMed]
- Bodart V, Bouchard JF, McNicoll N, et al. Identification and characterization of a new growth hormone-releasing peptide receptor in the heart. Circ Res 1999;85:796-802. [Crossref] [PubMed]
- Coraci IS, Husemann J, Berman JW, et al. CD36, a class B scavenger receptor, is expressed on microglia in Alzheimer's disease brains and can mediate production of reactive oxygen species in response to beta-amyloid fibrils. Am J Pathol 2002;160:101-12. [Crossref] [PubMed]
- Ren Y, Silverstein RL, Allen J, et al. CD36 gene transfer confers capacity for phagocytosis of cells undergoing apoptosis. J Exp Med 1995;181:1857-62. [Crossref] [PubMed]
- Ghosh A, Li W, Febbraio M, et al. Platelet CD36 mediates interactions with endothelial cell-derived microparticles and contributes to thrombosis in mice. J Clin Invest 2008;118:1934-43. [Crossref] [PubMed]
- Tandon NN, Kralisz U, Jamieson GA. Identification of glycoprotein IV (CD36) as a primary receptor for platelet-collagen adhesion. J Biol Chem 1989;264:7576-83. [Crossref] [PubMed]
- Puente Navazo MD, Daviet L, Ninio E, et al. Identification on human CD36 of a domain (155-183) implicated in binding oxidized low-density lipoproteins (Ox-LDL). Arterioscler Thromb Vasc Biol 1996;16:1033-9. [Crossref] [PubMed]
- Kar NS, Ashraf MZ, Valiyaveettil M, et al. Mapping and characterization of the binding site for specific oxidized phospholipids and oxidized low density lipoprotein of scavenger receptor CD36. J Biol Chem 2008;283:8765-71. [Crossref] [PubMed]
- Kuda O, Pietka TA, Demianova Z, et al. Sulfo-N-succinimidyl oleate (SSO) inhibits fatty acid uptake and signaling for intracellular calcium via binding CD36 lysine 164: SSO also inhibits oxidized low density lipoprotein uptake by macrophages. J Biol Chem 2013;288:15547-55. [Crossref] [PubMed]
- Cabrera A, Neculai D, Tran V, et al. Plasmodium falciparum-CD36 Structure-Function Relationships Defined by Ortholog Scanning Mutagenesis. J Infect Dis 2019;219:945-54. [Crossref] [PubMed]
- Navazo MD, Daviet L, Savill J, et al. Identification of a domain (155-183) on CD36 implicated in the phagocytosis of apoptotic neutrophils. J Biol Chem 1996;271:15381-5. [Crossref] [PubMed]
- Demers A, McNicoll N, Febbraio M, et al. Identification of the growth hormone-releasing peptide binding site in CD36: a photoaffinity cross-linking study. Biochem J 2004;382:417-24. [Crossref] [PubMed]
- Frieda S, Pearce A, Wu J, et al. Recombinant GST/CD36 fusion proteins define a thrombospondin binding domain. Evidence for a single calcium-dependent binding site on CD36. J Biol Chem 1995;270:2981-6. [Crossref] [PubMed]
- Crombie R, Silverstein R. Lysosomal integral membrane protein II binds thrombospondin-1. Structure-function homology with the cell adhesion molecule CD36 defines a conserved recognition motif. J Biol Chem 1998;273:4855-63. [Crossref] [PubMed]
- Tait JF, Smith C. Phosphatidylserine receptors: role of CD36 in binding of anionic phospholipid vesicles to monocytic cells. J Biol Chem 1999;274:3048-54. [Crossref] [PubMed]
- Yamaguchi A, Yamamoto N, Akamatsu N, et al. PS-liposome and ox-LDL bind to different sites of the immunodominant domain (#155-183) of CD36: a study with GS95, a new anti-CD36 monoclonal antibody. Thromb Res 2000;97:317-26. [Crossref] [PubMed]
- Doens D, Valiente PA, Mfuh AM, et al. Identification of Inhibitors of CD36-Amyloid Beta Binding as Potential Agents for Alzheimer's Disease. ACS Chem Neurosci 2017;8:1232-41. [Crossref] [PubMed]
- Luiken JJ, Chanda D, Nabben M, et al. Post-translational modifications of CD36 (SR-B2): Implications for regulation of myocellular fatty acid uptake. Biochim Biophys Acta 2016;1862:2253-8. [Crossref] [PubMed]
- Hoosdally SJ, Andress EJ, Wooding C, et al. The Human Scavenger Receptor CD36: glycosylation status and its role in trafficking and function. J Biol Chem 2009;284:16277-88. [Crossref] [PubMed]
- Kawecki C, Bocquet O, Schmelzer CEH, et al. Identification of CD36 as a new interaction partner of membrane NEU1: potential implication in the pro-atherogenic effects of the elastin receptor complex. Cell Mol Life Sci 2019;76:791-807. [Crossref] [PubMed]
- Hatmi M, Gavaret JM, Elalamy I, et al. Evidence for cAMP-dependent platelet ectoprotein kinase activity that phosphorylates platelet glycoprotein IV (CD36). J Biol Chem 1996;271:24776-80. [Crossref] [PubMed]
- Chu LY, Silverstein RL. CD36 ectodomain phosphorylation blocks thrombospondin-1 binding: structure-function relationships and regulation by protein kinase C. Arterioscler Thromb Vasc Biol 2012;32:760-7. [Crossref] [PubMed]
- Guthmann F, Maehl P, Preiss J, et al. Ectoprotein kinase-mediated phosphorylation of FAT/CD36 regulates palmitate uptake by human platelets. Cell Mol Life Sci 2002;59:1999-2003. [Crossref] [PubMed]
- Ho M, Hoang HL, Lee KM, et al. Ectophosphorylation of CD36 regulates cytoadherence of Plasmodium falciparum to microvascular endothelium under flow conditions. Infect Immun 2005;73:8179-87. [Crossref] [PubMed]
- Smith J, Su X, El-Maghrabi R, et al. Opposite regulation of CD36 ubiquitination by fatty acids and insulin: effects on fatty acid uptake. J Biol Chem 2008;283:13578-85. [Crossref] [PubMed]
- Xia X, Hu T, He J, et al. USP10 deletion inhibits macrophage-derived foam cell formation and cellular-oxidized low density lipoprotein uptake by promoting the degradation of CD36. Aging (Albany NY) 2020;12:22892-905. [Crossref] [PubMed]
- Zhang F, Xia X, Chai R, et al. Inhibition of USP14 suppresses the formation of foam cell by promoting CD36 degradation. J Cell Mol Med 2020;24:3292-302. [Crossref] [PubMed]
- Xia X, Xu Q, Liu M, et al. Deubiquitination of CD36 by UCHL1 promotes foam cell formation. Cell Death Dis 2020;11:636. [Crossref] [PubMed]
- Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [Crossref] [PubMed]
- Abumrad NA, Moore DJ. Parkin reinvents itself to regulate fatty acid metabolism by tagging CD36. J Clin Invest 2011;121:3389-92. [Crossref] [PubMed]
- Kim KY, Stevens MV, Akter MH, et al. Parkin is a lipid-responsive regulator of fat uptake in mice and mutant human cells. J Clin Invest 2011;121:3701-12. [Crossref] [PubMed]
- Lundby A, Lage K, Weinert BT, et al. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep 2012;2:419-31. [Crossref] [PubMed]
- Khan S, Kowluru A. CD36 mediates lipid accumulation in pancreatic beta cells under the duress of glucolipotoxic conditions: Novel roles of lysine deacetylases. Biochem Biophys Res Commun 2018;495:2221-6. [Crossref] [PubMed]
- Tao N, Wagner SJ, Lublin DM. CD36 is palmitoylated on both N- and C-terminal cytoplasmic tails. J Biol Chem 1996;271:22315-20. [Crossref] [PubMed]
- Thorne RF, Ralston KJ, de Bock CE, et al. Palmitoylation of CD36/FAT regulates the rate of its post-transcriptional processing in the endoplasmic reticulum. Biochim Biophys Acta 2010;1803:1298-307. [Crossref] [PubMed]
- van Oort MM, Drost R, Janßen L, et al. Each of the four intracellular cysteines of CD36 is essential for insulin- or AMP-activated protein kinase-induced CD36 translocation. Arch Physiol Biochem 2014;120:40-9. [Crossref] [PubMed]
- Hao JW, Wang J, Guo H, et al. CD36 facilitates fatty acid uptake by dynamic palmitoylation-regulated endocytosis. Nat Commun 2020;11:4765. [Crossref] [PubMed]
- Eyre NS, Cleland LG, Tandon NN, et al. Importance of the carboxyl terminus of FAT/CD36 for plasma membrane localization and function in long-chain fatty acid uptake. J Lipid Res 2007;48:528-42. [Crossref] [PubMed]
- McDermott-Roe C, Martin J, Collot-Teixeira S, et al. CD36 N-terminal cytoplasmic domain is not required for the internalization of oxidized low-density lipoprotein. Biosci Rep 2008;28:145-51. [Crossref] [PubMed]
- Rasmussen JT, Berglund L, Rasmussen MS, et al. Assignment of disulfide bridges in bovine CD36. Eur J Biochem 1998;257:488-94. [Crossref] [PubMed]
- Gruarin P, Sitia R, Alessio M. Formation of one or more intrachain disulphide bonds is required for the intracellular processing and transport of CD36. Biochem J 1997;328:635-42. [Crossref] [PubMed]
- Thorne RF, Meldrum CJ, Harris SJ, et al. CD36 forms covalently associated dimers and multimers in platelets and transfected COS-7 cells. Biochem Biophys Res Commun 1997;240:812-8. [Crossref] [PubMed]
- Jay AG, Chen AN, Paz MA, et al. CD36 binds oxidized low density lipoprotein (LDL) in a mechanism dependent upon fatty acid binding. J Biol Chem 2015;290:4590-603. [Crossref] [PubMed]
- Gruarin P, Primo L, Ferrandi C, et al. Cytoadherence of Plasmodium falciparum-infected erythrocytes is mediated by a redox-dependent conformational fraction of CD36. J Immunol 2001;167:6510-7. [Crossref] [PubMed]
- Wang R, Tao B, Fan Q, et al. Fatty-acid receptor CD36 functions as a hydrogen sulfide-targeted receptor with its Cys333-Cys272 disulfide bond serving as a specific molecular switch to accelerate gastric cancer metastasis. EBioMedicine 2019;45:108-23. [Crossref] [PubMed]
- Chadwick AC, Jensen DR, Hanson PJ, et al. NMR Structure of the C-Terminal Transmembrane Domain of the HDL Receptor, SR-BI, and a Functionally Relevant Leucine Zipper Motif. Structure 2017;25:446-57. [Crossref] [PubMed]
- Gaidukov L, Nager AR, Xu S, et al. Glycine dimerization motif in the N-terminal transmembrane domain of the high density lipoprotein receptor SR-BI required for normal receptor oligomerization and lipid transport. J Biol Chem 2011;286:18452-64. [Crossref] [PubMed]
- Marques PE, Nyegaard S, Collins RF, et al. Multimerization and Retention of the Scavenger Receptor SR-B1 in the Plasma Membrane. Dev Cell 2019;50:283-295.e5. [Crossref] [PubMed]
- Wei P, Sun FD, Zuo LM, et al. Critical residues and motifs for homodimerization of the first transmembrane domain of the plasma membrane glycoprotein CD36. J Biol Chem 2017;292:8683-93. [Crossref] [PubMed]
- Martin CA, Longman E, Wooding C, et al. Cd36, a class B scavenger receptor, functions as a monomer to bind acetylated and oxidized low-density lipoproteins. Protein Sci 2007;16:2531-41. [Crossref] [PubMed]
- Daviet L, Malvoisin E, Wild TF, et al. Thrombospondin induces dimerization of membrane-bound, but not soluble CD36. Thromb Haemost 1997;78:897-901. [Crossref] [PubMed]
- Triantafilou M, Gamper FG, Haston RM, et al. Membrane sorting of toll-like receptor (TLR)-2/6 and TLR2/1 heterodimers at the cell surface determines heterotypic associations with CD36 and intracellular targeting. J Biol Chem 2006;281:31002-11. [Crossref] [PubMed]
- Stewart CR, Stuart LM, Wilkinson K, et al. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat Immunol 2010;11:155-61. [Crossref] [PubMed]
- Kennedy DJ, Chen Y, Huang W, et al. CD36 and Na/K-ATPase-α1 form a proinflammatory signaling loop in kidney. Hypertension 2013;61:216-24. [Crossref] [PubMed]
- Miao WM, Vasile E, Lane WS, et al. CD36 associates with CD9 and integrins on human blood platelets. Blood 2001;97:1689-96. [Crossref] [PubMed]
- Huang W, Febbraio M, Silverstein RL. CD9 tetraspanin interacts with CD36 on the surface of macrophages: a possible regulatory influence on uptake of oxidized low density lipoprotein. PLoS One 2011;6:e29092. [Crossref] [PubMed]
- Kazerounian S, Duquette M, Reyes MA, et al. Priming of the vascular endothelial growth factor signaling pathway by thrombospondin-1, CD36, and spleen tyrosine kinase. Blood 2011;117:4658-66. [Crossref] [PubMed]
- Bamberger ME, Harris ME, McDonald DR, et al. A cell surface receptor complex for fibrillar beta-amyloid mediates microglial activation. J Neurosci 2003;23:2665-74. [Crossref] [PubMed]
- Heit B, Kim H, Cosío G, et al. Multimolecular signaling complexes enable Syk-mediated signaling of CD36 internalization. Dev Cell 2013;24:372-83. [Crossref] [PubMed]
- Huang MM, Bolen JB, Barnwell JW, et al. Membrane glycoprotein IV (CD36) is physically associated with the Fyn, Lyn, and Yes protein-tyrosine kinases in human platelets. Proc Natl Acad Sci U S A 1991;88:7844-8. [Crossref] [PubMed]
- Yang M, Li W, Harberg C, et al. Cysteine sulfenylation by CD36 signaling promotes arterial thrombosis in dyslipidemia. Blood Adv 2020;4:4494-507. [Crossref] [PubMed]
- Stuart LM, Deng J, Silver JM, et al. Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J Cell Biol 2005;170:477-85. [Crossref] [PubMed]
- Nergiz-Unal R, Lamers MM, Van Kruchten R, et al. Signaling role of CD36 in platelet activation and thrombus formation on immobilized thrombospondin or oxidized low-density lipoprotein. J Thromb Haemost 2011;9:1835-46. [Crossref] [PubMed]
Cite this article as: May SC, Sahoo D. Now in 3D! Novel insights into CD36 structure and function. Ann Blood 2021;6:33.


